Publications
At UW-Madison:
- Buck, K. M.; Rogers, H. T.; Gregorich, Z. R.; Mann, M. W.; Aballo, T. J.; Gao, Z.; Chapman, E. A.; Perciaccante, A. J.; Price, S. J.; Lei, I.; Tang, P. C.; Ge, Y. Extracellular matrix alterations in chronic ischemic cardiomyopathy revealed by quantitative proteomics. JCI Insight 2025. PrePrint
- Raza, F.; Gregorich, Z. R.; Freeman, J.; Houston, T.; Garcia-Arango, M.; Lechuga, C.; Chen, Y.; Sahai, A.; Shaer, A. E.; Korcarz, C.; Cui, K.; Park, Y.; Jones, K.; Tu, W.; Runo, J.; Schulte, J. J.; Nagpal, P.; Ge, Y.; Chesler, N.; Stewart, R.; Wieben, O.; Guo, W., Multimodal Characterization of High-risk PH-HFpEF phenogroup with Right Ventricular Dysfunction: Vascular Mechanics and Myocardial Transcriptomics. medRxiv 2025. Article
- Wilson, M. C.; Josvai, M.; Walters, J. K.; Lawson, J.; Rossler, K. J.; Gao, Z.; Zhu, Y.; Kamp, T. J.; Crone, W. C.; Eckhardt, L. L.; Ge, Y., High-Sensitivity Top-Down Proteomics Reveals Enhanced Maturation of Micropatterned Induced Pluripotent Stem Cell-Derived Cardiomyocytes. J. Proteome Res. 2025. Article
- Lei, I.; Sicim, H.; Gao, W.; Huang, W.; Noly, P. E.; Pergande, M. R.; Wilson, M. C.; Lee, A.; Liu, L.; Abou El Ela, A.; Jiang, M.; Saddoughi, S. A.; Pober, J. S.; Platt, J. L.; Cascalho, M.; Pagani, F. D.; Chen, Y. E.; Pitt, B.; Wang, Z.; Mortensen, R. M.; Ge, Y.; Tang, P. C. Mineralocorticoid receptor phase separation modulates cardiac preservation. Nat. Cardiovasc. Res. 2025, 4, 710–726. Article
- Chan, H.-J.; Krichel, B.; Bandura, L. J.; Chapman, E. A.; Rogers, H. T.; Fischer, M. S.; Roberts, D. S.; Gao, Z.; Wang, M.-D.; Wu, J.; Uetrecht, C.; Jin, S.; Ge, Y. Structural heterogeneity of proteoform-ligand complexes in amp-activated protein kinase uncovered by integrated top-down mass spectrometry. J. Am. Chem. Soc. 2025, 147, 30809–30819 Article
- Fischer, M. S.; Rogers, H. T.; Chapman, E. A.; Jin, S.; Ge, Y., Native top-down proteomics of endogenous protein complexes enabled by online two-dimensional liquid chromatography. Anal. Chem. 2025, 97, 25, 13663-13671 Article
- Liu, M.; Zhu, Y.; McIlwain, S. J.; Deng, H.; Brasier, A. R.; Ge, Y.; Kimple, M. E.; Baschnagel, A. M., Characterizing Plasma-based Metabolomic Signatures for Metastasis in Non- Small Cell Lung Cancer. Metabolites. 2025. Article
- Liu, X.; Roberts, D. S.; Bingman, C. A.; Jin, S.; Ge, Y.; Gellman, S. H., Rational design of a foldon-derived heterotrimer guided by quantitative native mass spectrometry. JACS 2025, 2025.03. 25.645370. Article
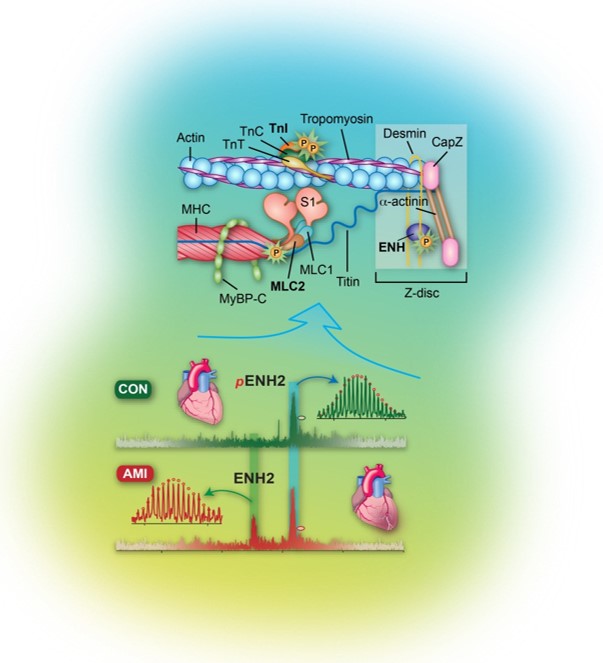
- Aballo, T. J.; Bae, J.; Paltzer, W. G.; Chapman, E. A.; Perciaccante, A. J.; Pergande, M. R.; Salamon, R. J.; Nuttall, D. J.; Mann, M. W.; Ge, Y., Integrated Proteomics identifies troponin I isoform switch as a Regulator of a Sarcomere-Metabolism Axis during Cardiac Regeneration. Cardiovasc. Res. 2025, cvaf069. Article

- Towler, A. G.; Perciaccante, A. J.; Aballo, T. J.; Zhu, Y.; Wang, F.; Lloyd, S.; Kadoya, K.; He, Y.; Tian, Y.; Ge, Y., A Single-Step Protein Extraction for Lung Extracellular Matrix Proteomics Enabled by the Photocleavable Surfactant Azo and timsTOF Pro. Mol. Cell.Proteomics 2025, 100950. Article

- Reasoner, E. A.; Chan, H.-J.; Aballo, T. J.; Plouff, K. J.; Noh, S.; Ge, Y.; Jin, S., In Situ Metal–Organic Framework Growth in Serum Encapsulates and Depletes Abundant Proteins for Integrated Plasma Proteomics. ACS nano 2025, 19, 13968–13981. Article

- Knott, S. J.; Tucholski, T.; Josyer, H.; Inman, D.; Friedl, A.; Zhu, Y.;; Ge Y.; Ponik, S. M., Deciphering Proteoform Landscape of Mammary Carcinoma by Top-Down Proteomics. J. Proteome Res. 2025, 24, 3, 1425–1438. Article

- Chapman, E. A.; Rogers, H. T.; Gao, Z.; Chan, H.-J.; Alvarado, F. J.; Ge, Y., In-depth characterization of S-glutathionylation in ventricular myosin light chain 1 across species by top-down proteomics. J. Mol. Cell. Cardiol. 2025. doi: 10.1016/j.yjmcc.2025.03.012. Online ahead of print. Article

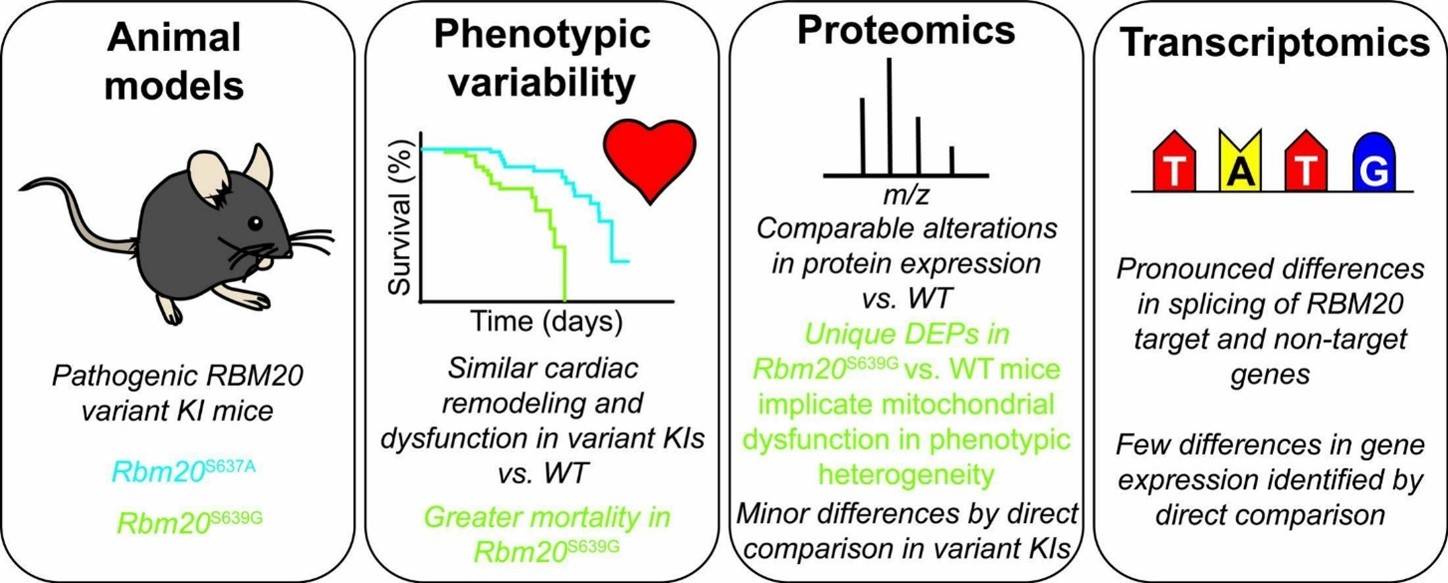
- Gregorich, Z.R.; Larson, E.J.; Zhang, Y.; Braz, C.U.; Ge, Y.; Guo, W., Integrated
proteomics and transcriptomics analysis reveals insights into differences in premature mortality
associated with disparate pathogenic RBM20 variants. J. Mol. Cell. Cardiol., 2024,
197, 78-89. Article
- Turner, D.G.P.; De Lange, W. J.; Zhu, Y.; Coe, C.L.; Simcox, J.; Ge, Y.;
Kamp, T.J.; Ralphe, J.C.; Glukhov, A.V., Neutral sphingomyelinase regulates mechanotransduction in
human engineered cardiac tissues and mouse hearts. J. Physiol., 2024,
602 (18), 4387-4407 Article
- Fischer, M.S.; Rogers, H.T.; Chapman, E.A.; Chan, H.; Krichel, B.; Gao, Z.;
Larson, E.J.; Ge, Y., Online Mixed-Bed Ion Exchange Chromatography for
Native Top-Down Proteomics of Complex Mixtures. J. Proteome Res., 2024,
23 (7), 2315-2322. Article
- Roberts, D. S.; Loo, J. A.; Tsybin, Y. O.; Liu, X.; Wu, S.; Chamot-Rooke, J.;
Agar, J. N.; Paša-Tolić, L.; Smith, L. M.; Ge, Y., Top-down proteomics. Nature Reviews Methods
Primers 2024, 4 (1), 38. Article

- Wang, Y.; Zhao, M.; Liu, X.; Xu, B.; Reddy, G. R.; Jovanovic, A.; Wang, Q.; Zhu, C.;
Xu, H.; Bayne, E. F.; Xiang, W.; Tilley, D. G.; Ge, Y.; Tate, C. G.; Feil, R.; Chiu, J. C.; Bers, D. M.;
Xiang, Y. K., Carvedilol Activates a Myofilament Signaling Circuitry to Restore Cardiac Contractility in
Heart Failure. JACC: Basic to Translational Science 2024. Article

- Habeck, T.; Brown, K. A.; Des Soye, B.; Lantz, C.; Zhou, M.; Alam, N.; Hossain, M. A.; Jung, W.; Keener, J. E.; Volny, M.; Wilson, J. W.; Ying, Y.; Agar, J. N.; Danis, P. O.; Ge, Y.; Kelleher, N. L.; Li, H.; Loo, J. A.; Marty, M. T.; Paša-Tolić, L.; Sandoval, W.; Lermyte, F., Top-down mass spectrometry of native proteoforms and their complexes: a community study. Nature Methods 2024. Article
- Stroik, D.; Gregorich, Z. R.; Raza, F.; Ge, Y.; Guo, W., Titin: roles in cardiac
function and diseases. Front Physiol 2024, 15, 1385821. Article

- Anderson, C. L.; Brown, K. A.; North, R. J.; Walters, J. K.; Kaska, S. T.;
Wolff, M. R.; Kamp, T. J.; Ge, Y.; Eckhardt, L. L., Global Proteomic Analysis Reveals Alterations in
Differentially Expressed Proteins between Cardiopathic Lamin A/C Mutations. Journal of Proteome Research
2024,23 (6), 1970-1982. Article

- Wancewicz, B.; Pergande, M. R.; Zhu, Y.; Gao, Z.; Shi, Z.; Plouff, K.; Ge, Y.,
Comprehensive Metabolomic Analysis of Human Heart Tissue Enabled by Parallel Metabolite Extraction
and High-Resolution Mass Spectrometry. Anal Chem 2024,96 (15), 5781-5789. Article

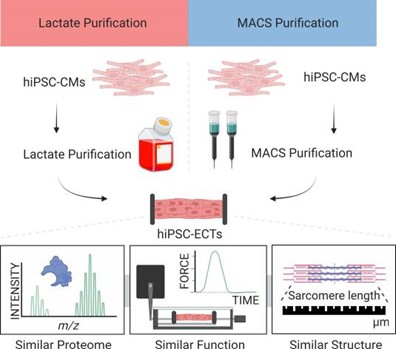
- Rossler, K. J.; de Lange, W. J.; Mann, M. W.; Aballo, T. J.; Melby, J. A.; Zhang, J.; Kim,
G.; Bayne, E. F.; Zhu, Y.; Farrell, E. T.; Kamp, T. J.; Ralphe, J. C.; Ge, Y., Lactate- and
immunomagnetic-purified hiPSC-derived cardiomyocytes generate comparable engineered
cardiac tissue constructs. JCI Insight, 2024, Article
- Chapman, E. A.; Li, B. H.; Krichel, B.; Chan, H.-J.; Buck, K. M.; Roberts, D. S.; Ge, Y.
Native Top-Down Mass Spectrometry for Characterizing Sarcomeric Proteins Directly from
Cardiac Tissue Lysate. J. Am. Soc. Spectr. 2024, Article
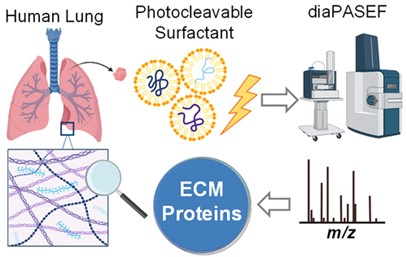
- Bayne, E. F.; Buck, K. M.; Tower, A. G.; Zhu, Y.; Pergande, M. R.; Zhou, T.; Price, S.;
Rossler, K. J.; Morales-Tirado, V.; Lloyd, S.; Wang, F.; He, Y.; Tian, Y.; Ge, Y. High-Throughput
Extracellular Matrix Proteomics of Human Lungs Enabled by Photocleavable Surfactant and
diaPASEF. J Proteome Res 2024, Article
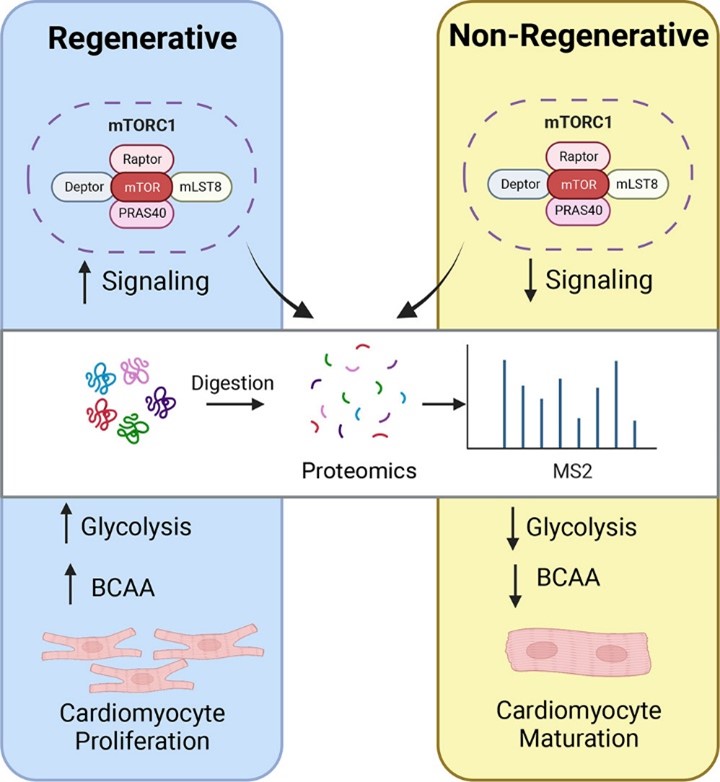
- Paltzer, W. G.; Aballo, T. J.; Bae, J.; Flynn, C. G. K.; Wanless, K. N.; Hubert, K. A.;
Nuttall, D. J.; Perry, C.; Malawi, R.; Ge, Y.; Mahmoud, A. I., mTORC1 regulates the metabolic
switch of postnatal cardiomyocytes during regeneration. J. Mol. Cell. Cardiol. 2024 Article
- Rogers, H. T.; Melby, J. A.; Ehlers, L. E.; Fischer, M. S.; Larson, E. J.; Gao, Z.; Rossler, K.
J.; Wang, D.; Alpert, A. J.; Ge, Y. Small-Scale Serial Size Exclusion Chromatography (s3SEC)
for High Sensitivity Top-Down Proteomics of Large Proteoforms. Anal. Chem. 2024 Article
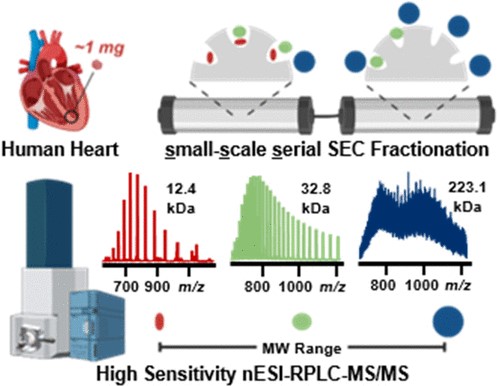
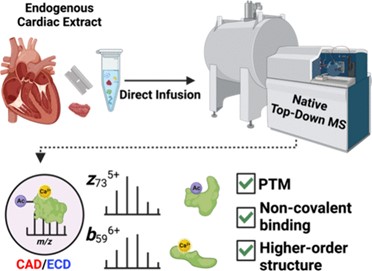
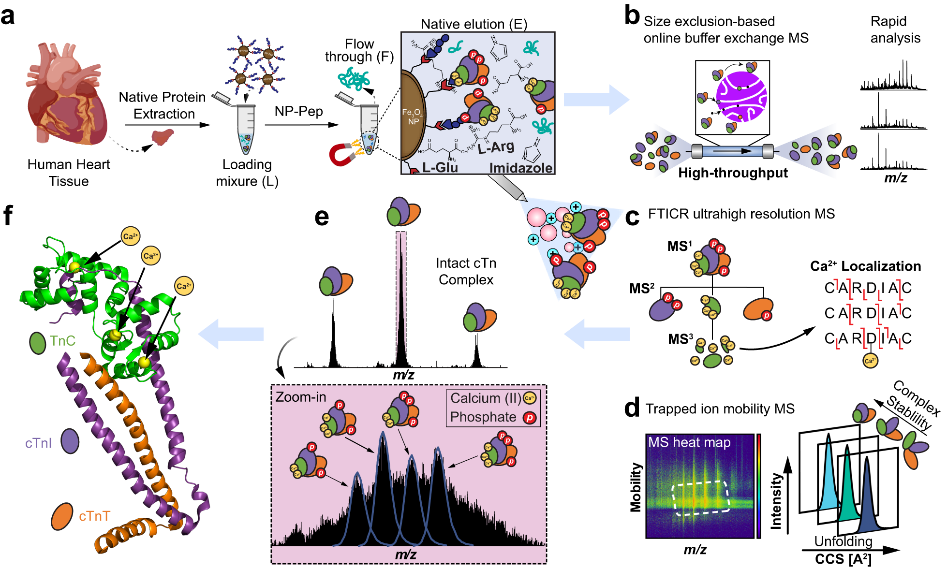
- Chapman, E. A.; Roberts, D. S.; Tiambeng, T. N.; Andrews, J.; Wang, M.-D.; Reasoner, E. A.; Melby, J. A.; Li, B. H.; Kim, D.; Alpert, A. J.; Jin, S.; Ge, Y. Structure and dynamics of endogenous cardiac troponin complex in human heart tissue captured by native nanoproteomics. Nature Communications 2023, 14, 8400. Article
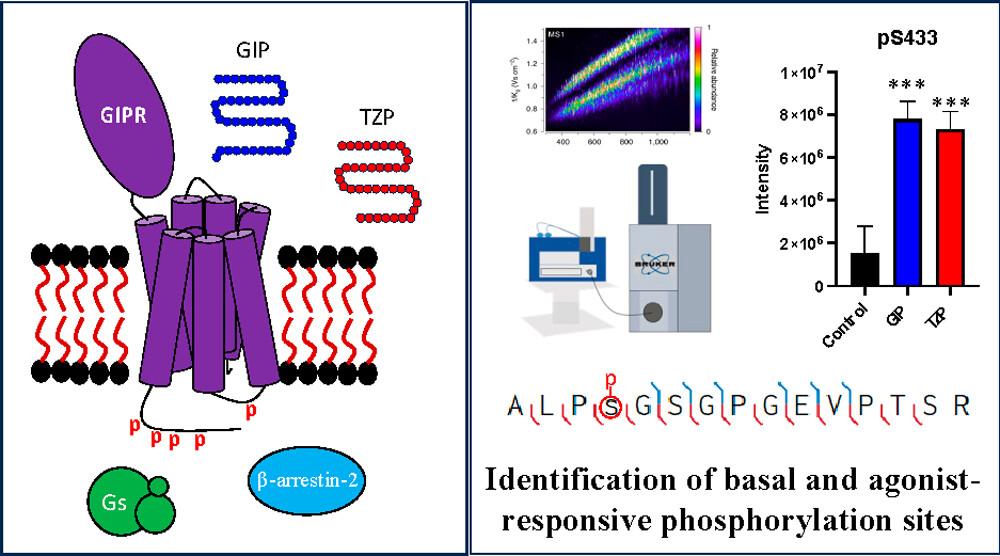
- Brown, K. A.; Morris, R. K.; Eckhardt, S. J.; Ge, Y.; Gellman, S. H. Phosphorylation Sites of the Gastric Inhibitory Polypeptide Receptor (GIPR) Revealed by Trapped-Ion-Mobility Spectrometry Coupled to Time-of-Flight Mass Spectrometry (TIMS-TOF MS). J. Am. Chem. Soc. 2023, Epub ahead of Print Article
- West, K. H. J.; Ma, S. V.; Pensinger, D. A.; Tucholski, T.; Tiambeng, T. N.; Eisenbraun, E. L.; Yehuda, A.; Hayouka, Z.; Ge, Y.; Sauer, J.; Blackwell, H. E. Characterization of an Autoinducing Peptide Signal Reveals Highly Efficacious Synthetic Inhibitors and Activators of Quorum Sensing and Biofilm Formation in Listeria monocytogenes. Biochemistry. 2023. Article
- Lermyte, F.; Habeck, T.; Brown, K.; Des Soye, B.; Lantz, C.; Zhou, M.; Alam, N.; Hossain, M.A.; Jung, W.; Keener, J.; Volny, M.; Wilson, J.; Ying, Y.; Agar, J.; Danis, P.; Ge, Y.; Kelleher, N.; Li, H.; Loo, J.; Marty, M.; Pasa-Tolic, L.; Sandoval, W. Top-down mass spectrometry of native proteoforms and their complexes: A community study. Under review at Nature Portfolio. 2023. PREPRINT
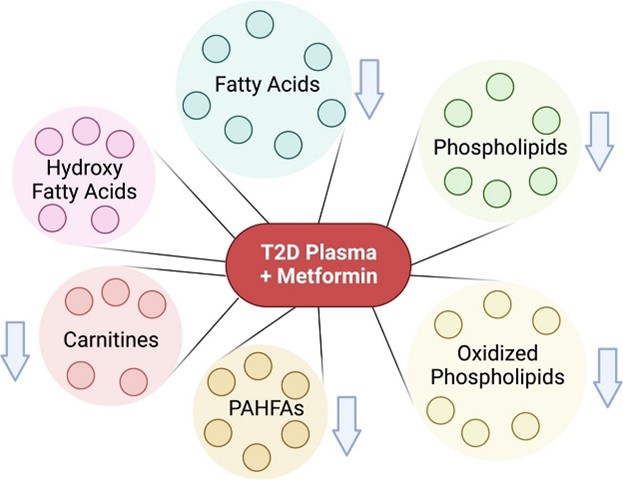
- Wancewicz, B.; Zhu, Y.; Fenske, R.J.; Weeks, A.M.; Wenger, K.; Pabich, S.; Daniels, M.; Punt, M.; Nall, R.; Peter, D.C.; Brasier, A.; Cox, E.D.; Davis, D.B.; Ge, Y.; Kimple, M.E. Metformin monotherapy alters the human plasma lipidome independent of clinical markers of glycemic control and cardiovascular disease risk in a type 2 diabetes clinical cohort. Journal of Pharmacology and Experimental Therapeutics. 2023. Article
- Yan, J.; Wu, Q.; Maity, M.; Braun, D.R.; Alas, I.; Wang, X.; Ying, X.; Zhu, Y.; Bell, B.A.; Rajski, S.R.; Ge, Y.; Richardson, D.D.; Zhong, W.; Bugni, T.S. Rapid Unambiguous Structure Elucidation of Streptnatamide A, a New Cyclic Peptide Isolated from A Marine-derived Streptomyces sp. Chemistry -A European Journal. 2023. Article
- Pergande, M. R.; Osterbauer, K. J.; Buck, K. M.; Roberts, D. S.; Wood, N. N.; Balasubramanian, P.; Mann, M. W.; Rossler, K. J.; Diffee, G. M.; Colman, R. J.; Anderson, R. M.; Ge, Y. Mass Spectrometry-Based Multiomics Identifies Metabolic Signatures of Sarcopenia in Rhesus Monkey Skeletal Muscle. Journal of Proteome Research 2023.Article
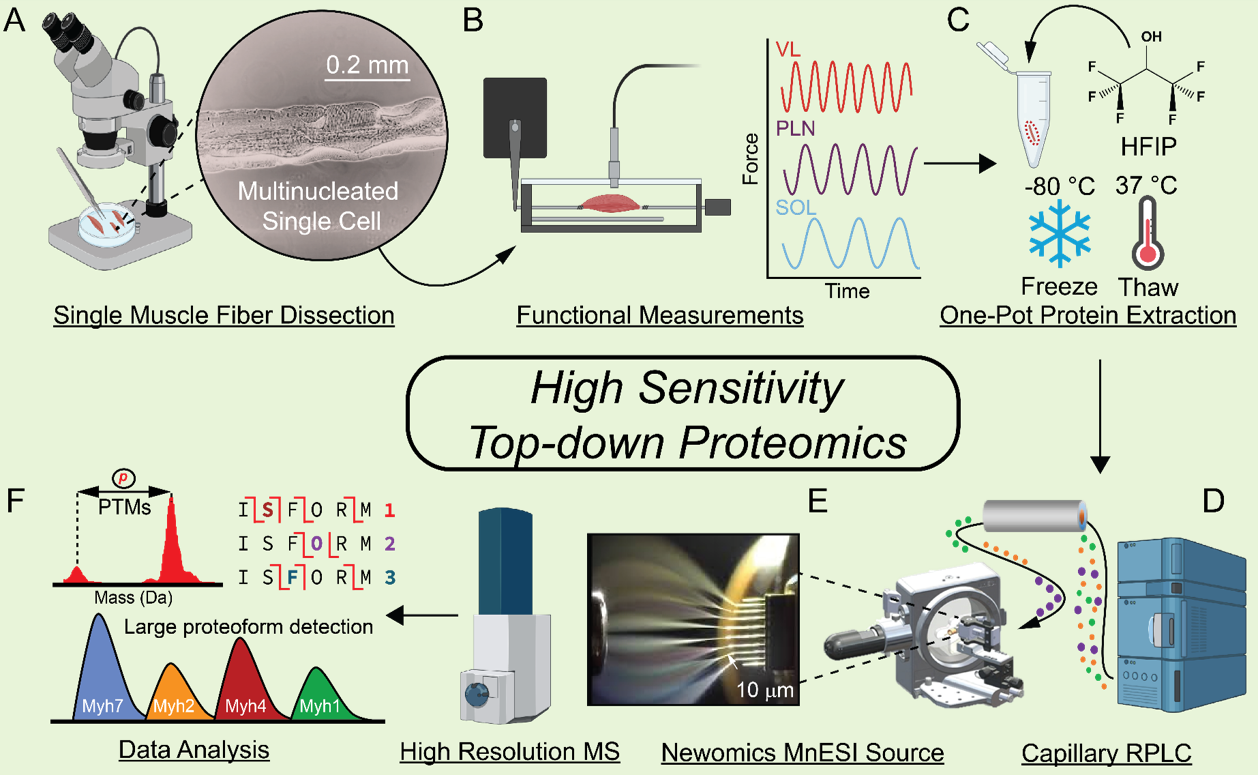
- Melby, J.A.; Brown, K.A.; Gregorich, Z.R.; Roberts, D.S.; Chapman, E.A.; Ehlers, L.E.; Gao, Z.; Larson, E.J.; Jin, Y.; Lopez, J.R.; Hartung, J.; Zhu, Y.; McIlwain, S.J.; Wang, D.; Guo, W.; Diffee, G.M.; Ge, Y. High sensitivity top-down proteomics captures single muscle cell heterogeneity in large proteoforms. PNAS. 2023, 120(19), e2222081120. Article
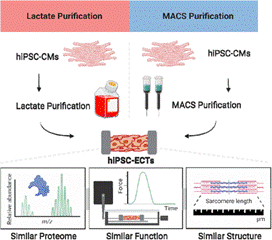
- Rossler, K.J.; de Lange, W.; Mann, M.W.; Aballo, T.J.; Melby, J.A.; Zhang, J.; Kim, G.; Bayne, E.F.; Zhu, Y.; Farrell, E.T.; Kamp, T.J.; Ralphe, J.C.; Ge, Y. Lactate and Immunomagnetic-purified iPSC-derived Cardiomyocytes Generate Comparable Engineered Cardiac Tissue Constructs. JCI Insight. 2023. Article
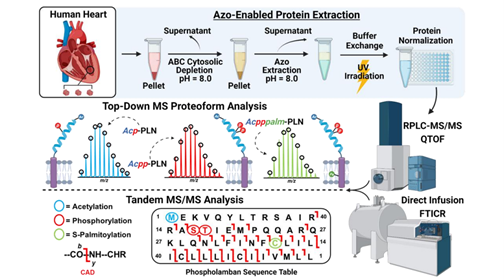
- Rogers, H.T.; Roberts, D.S.; Larson, E.J.; Melby, J.A.; Rossler, K.J.; Carr, A.V.; Brown, K.A.; Ge, Y. Comprehensive Characterization of Endogenous Phospholamban Proteoforms Enabled by Photocleavable Surfactant and Top-down Proteomics. Anal. Chem. 2023. Article
- Wancewicz, B.; Zhu, Y.; Fenske, R.K.; Weeks, A.M.; Wenger, K.; Pabich, S.; Daniels, M.; Punt, M.; Nall, R.; Peter, D.C.; Brasier, A.; Cox, E.D.; Davis, D.B.; Ge, Y.; Kimple, M. Metformin Monotherapy Alters the Human Plasma Lipidome Independent of Clinical Markers of Glycemic Control and Cardiovascular Disease Risk in a Type 2 Diabetes Clinical Cohort. J. of Pharmacology and Exp. Therapeutics. 2023. Article
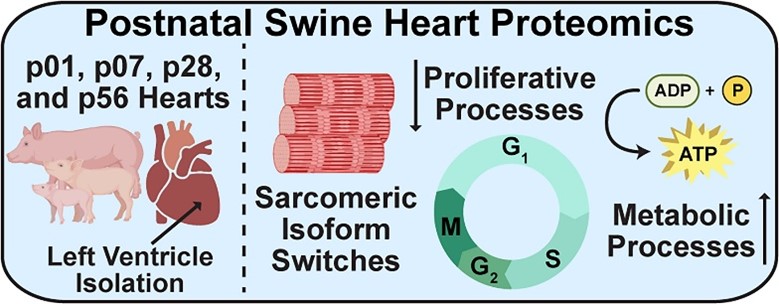
- Aballo, T. J.; Roberts, D.S.; Bayne, E. F.; Zhu, W.; Walcott, G.; Mahmoud, A. I.; Zhang, J.; Ge, Y. Integrated Proteomics Reveals Alterations in Sarcomere Composition and Developmental Processes during Postnatal Swine Heart Development, J. Mol. Cell. Cardiol. 2023, 176, 33-40 Article
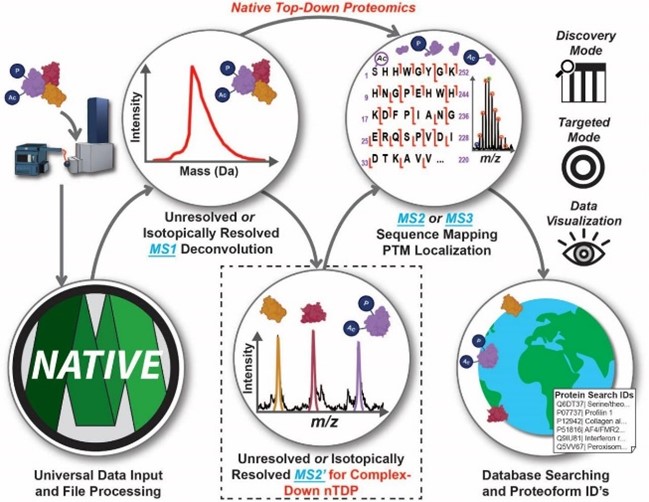
- Larson, E.J. ; Pergande, M. R; Moss, M. E.; Rossler, K. J.; Wenger, R. K.; Krichel, B.; Josyer, H.; Melby, J. A.; Roberts, D. S.; Pike, K.; Shi, Z.; Chan, H.; Knight, B.; Rogers, H. T.; Brown, K. A.; Ong, I. M.; Kyowon Jeong, K.; Marty, M.; McIlwain, S. J.; Ge, Y. MASH Native: A Unified Solution for Native Top-Down Proteomics Data Processing Bioinformatics 2023, btad359 Article
- Mann, M.; Fu, Y.; Xu, X.; Roberts, D.S.; Li, Y.; Zhou, J., Ge, Y.; Brasier, A.R. Bromodomain-containing Protein 4 Regulates Innate Inflammation in Airway Epithelial Cells via Modulation of Alternative Splicing. Front. Immunol., 2023, 14, Article
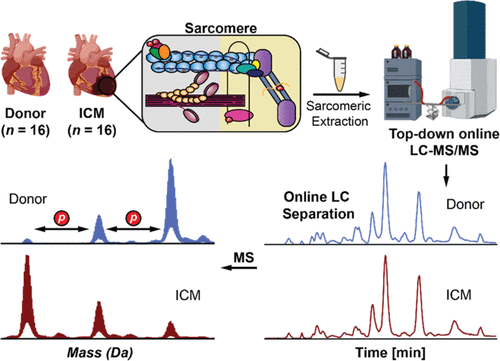
- Chapman, E.; Aballo, J. J.; Melby, J.A.; Zhou, T.; Price, S. J.; Rossler, K. J.; Lei, I.; Tang, P. C.; Ge, Y. Defining the Sarcomeric Proteoform Landscape in Ischemic Cardiomyopathy by Top-down Proteomics, J. Proteome Res. 2023, 22, 931-941 Article
- Bayne, E.F.; Rossler, K. J.; Gregorich, Z. R.; Aballo, T. J.; Roberts, D.S.; Chapman, E. A.;Guo, W.; Ralphe, J.C.; Kamp, T.J.; Ge, Y. Top-down Proteomics of Myosin Light Chain Isoforms Define Chamber-Specific Expression in the Human Heart, J Mol Cell Cardiol 2023, 181, 89-97. Article
- Brown, K. A.; Gugger, M. K.; Roberts, D. S.; Moreno, M.; Chae, P. S.; Ge, Y.; Jin, S. Synthesis, Self-Assembly Properties, and Degradation Characterization of a Nonionic Photocleavable Azo-Sulfide Surfactant Family, Langmuir, 2023, DOI: 10.1021/acs.langmuir.2c02820 Article
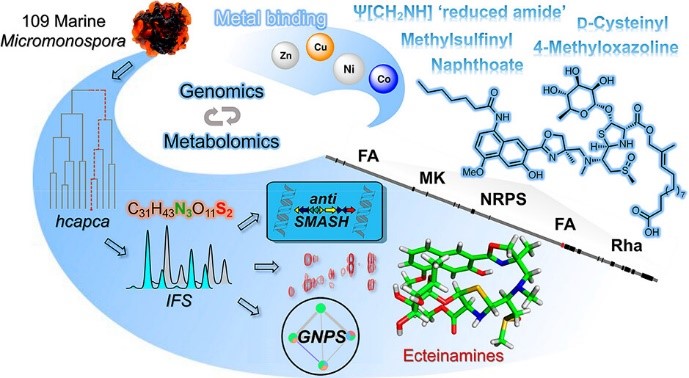
- Wu, Q.; Bell, B.A.; Yan, J.; Chevrette, M.G.; Brittin, N.J.; Zhu, Y.; Chanana, S.; Maity, M.; Braun, D.R.; Wheaton, A.M.; Guzei, I.A.; Ge, Y.; Rajski, S.R.; Thomas, M.G.; Bugni, T.S.; SR Metabolomics and Genomics Enable the Discovery of a New Class of Nonribosomal Peptidic Metallophores from a Marine Micromonospora. J. Am. Chem. Soc. 2023, 145, 58-69. Article
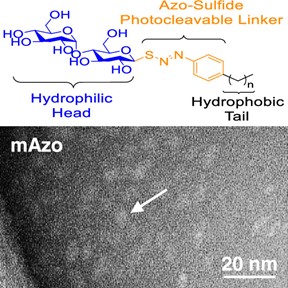
- Brown, K.A.; Gugger, M.; Yu, Z.; Moreno, D.; Jin, S.; Ge, Y. A Nonionic, Cleavable Surfactant for Native Mass Spectrometry and Top-down Proteomics, Anal. Chem. 2023, 95, 1801-1804 Article
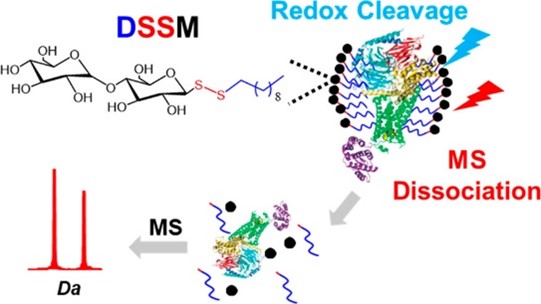
- Wu, Q.; Bell, B.A.; Yan, J.; Chevrette, M.G.; Brittin, N.J.; Zhu, Y.; Chanana, S.; Maity, M.; Braun, D.R.; Wheaton, A.M.; Guzei, I.A.; Ge, Y.; Rajski, S.R.; Thomas, M.G.; Bugni, T.S.; SR Metabolomics and Genomics Enable the Discovery of a New Class of Nonribosomal Peptidic Metallophores from a Marine Micromonospora. J. Am. Chem. Soc. 2023, 145, 58-69. Article
- Sun M.; Jin Y.; Zhang Y.; Gregorich Z.R.; Ren J.; Ge Y.; Guo W.; SR Protein Kinases Regulate the Splicing of Cardiomyopathy-Relevant Genes via Phosphorylation of the RSRSP Stretch in RBM20. Genes, 2022, 13(9), 1526. Article.
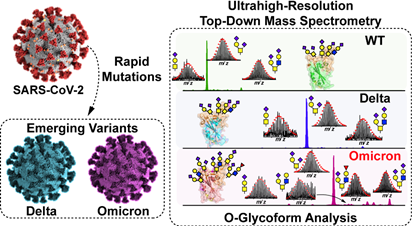
- Roberts, D.S.; Mann, M.; Li, B.H.; Kim, D.; Brasier, A.R.; Jin, S. Ge, Y. Distinct Core Glycan and O-Glycoform Utilization of SARS-CoV-2 Omicron Variant Spike Protein RBD Revealed by Top-Down Mass Spectrometry. Chem. Sci. 2022, 13, 10944-10949. Article.
- Larson, E.J.; Gregorich, Z.R.; Zhang, Y.; Li, B.H.; Aballo, T.J.; Melby, J.A.; Ge, Y.; Guo, W. RBM20 Ablation is Associated With Changes in The Expression of Titin-Interacting and Metabolic Proteins. Molecular Omics, 2022. Article.
- Gupta, K.; Brown, K.A.; Hsieh, M.L.; Hoover, B.; Wang, J.; Khoury, M.K.; Pilli, V.S.S.; Beyer, R.S.; Voruganti, N.R.; Chaudhary, S.; Roberts, D.S.; Murphy, R.M.; Hong, S.; Ge, Y.; Liu, B. Necroptosis is associated with Rab27-independent expulsion of extracellular vesicles containing RIPK3 and MLKL. Journal of Extracellular Vesicles, 2022, 11, e12261. Article.
- Zhu, Y.; Esnault, S.; Ge, Y.; Jarjour, N.N; Brasier, A.R. Airway fibrin formation cascade in allergic asthma exacerbation: implications for inflammation and remodeling. Clin Proteomics. 2022, 19(1), 15. Article.
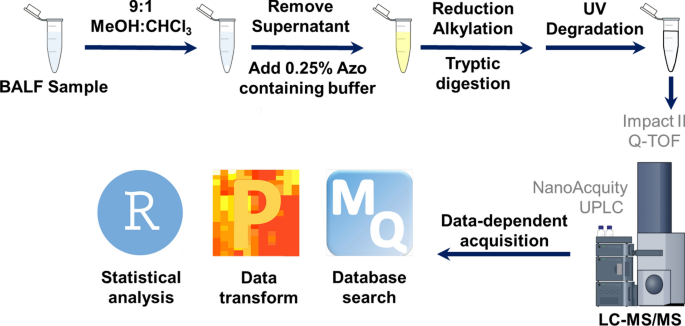
- Zhu, Y.; Esnault, S.; Ge, Y.; Jarjour, N.N.; Brasier, A.R. Segmental Bronchial Allergen Challenge Elicits Distinct Metabolic Phenotypes in Allergic Asthma. Metabolites, 2022, 12(5), 381. Article.
- Tiambeng, T.N.; Wu, Z.; Melby, J.A.; Ge, Y. Size Exclusion Chromatography Strategies and MASH Explorer for Large Proteoform Characterization. Methods Mol Biol. 2022, 2500, 15-30. Article.
- Zhang, Y.; Wang, C.; Sun, M.; Jin, Y.; Braz, C.U.; Khatib, H.; Hacker, T.A.; Liss, M.; Gotthardt, M.; Granzier, H.; Ge, Y.; Guo, W. RBM20 phosphorylation and its role in nucleocytoplasmic transport and cardiac pathogenesis. FASEB Journal, 2022, Epub ahead of print. Article.
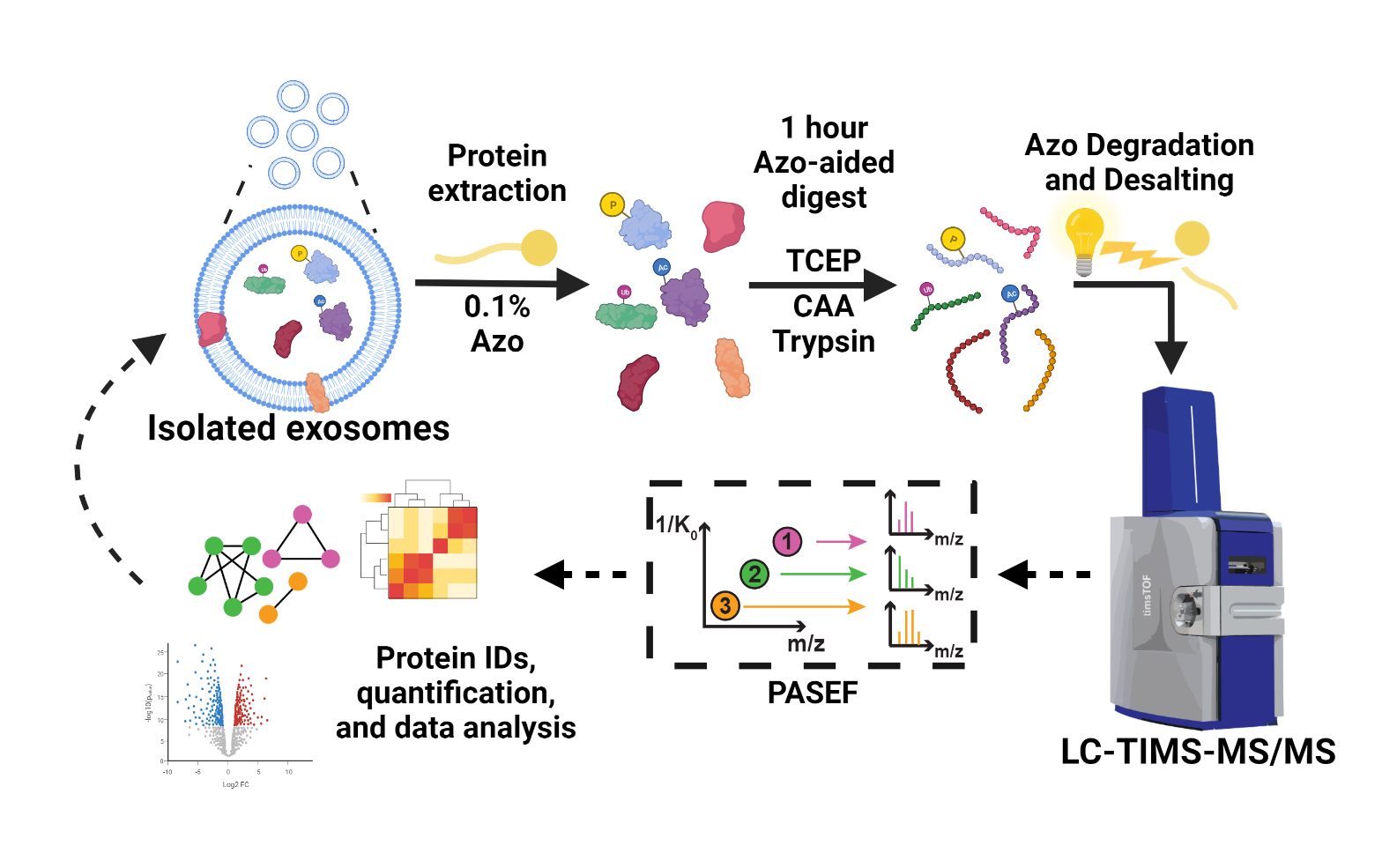
- Buck, K.M.; Roberts, D.S.; Aballo, T.J.; Inman, D.R.; Jin, S.; Ponik, S.; Brown, K.A.; Ge, Y. One-Pot Exosome Proteomics Enabled by a Photocleavable Surfactant. Anal. Chem., 2022, 94, 7164-7168. Article.
- Li, A.; Campbell, K.; Lal, S.; Ge, Y.; Keogh, A.; Macdonald, P.S.; Lau, P.; Linke, W.A.; Van der Velden, J.; Field, A.; Martinac, B.; Grosser, M.; dos Remedios, C. Peripartum cardiomyopathy: a global effort to find the cause and cure for the rare and little understood disease Biophysical Reviews. 2022, 14, 369-379. Article.
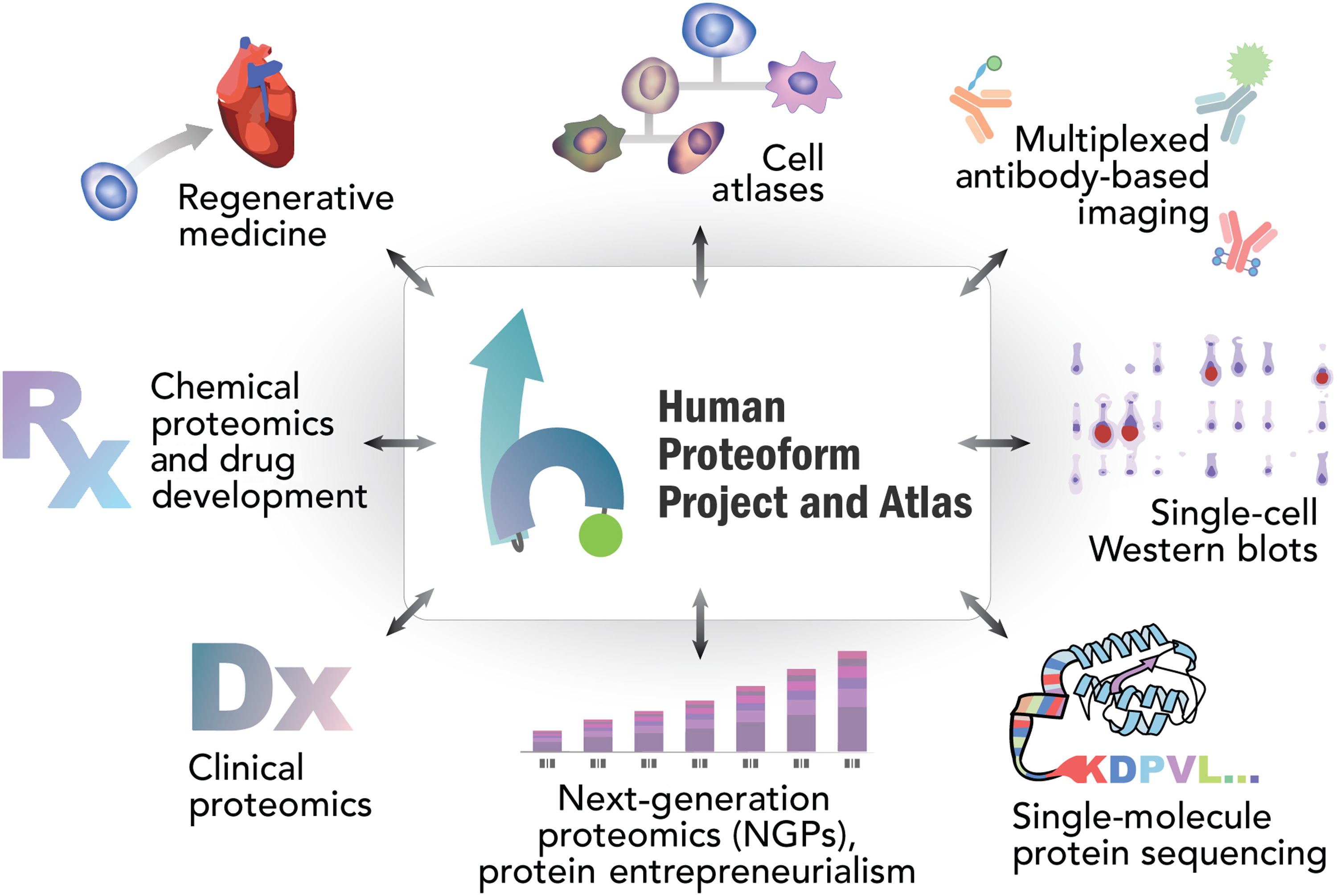
- Smith, L.M.; Agar, J.N.; Chamot-Rooke, J.; Danis, P.O.; Ge, Y. ; Loo, J.A.; Pasa-Tolic, L.; Tsybin, Y.O.; Kelleher, N.L. The Human Proteoform Project: Defining the Human Proteome Science Adv., 2021, 7, 46. Article.
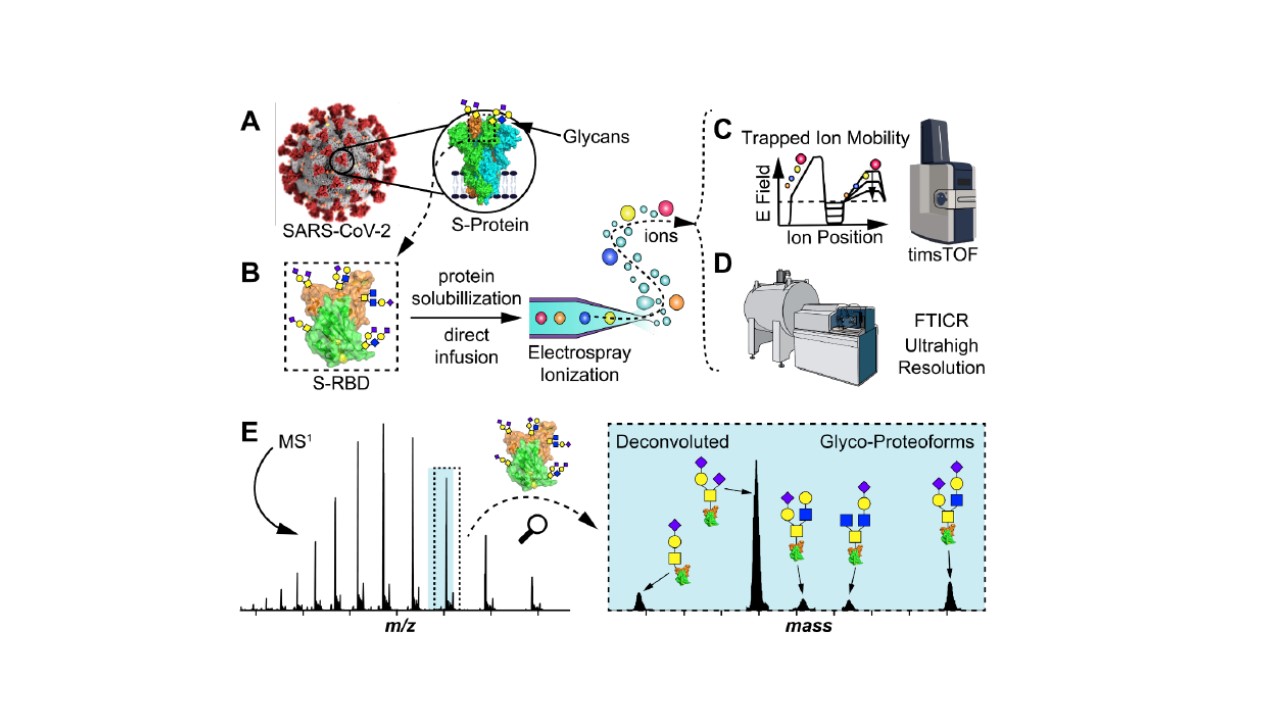
- Roberts, D.S.; Mann, M.; Melby, J.A.; Larson, E.J.; Zhu, Y.; Brasier, A.R.; Jin, S.; Ge, Y. Structural O-Glycoform Heterogeneity of the SARS-CoV-2 Spike Protein Receptor-Binding Domain Revealed by Native Top-Down Mass Spectrometry J. Am. Chem. Soc., 2021, 143, 31, 12014-12024. Article.
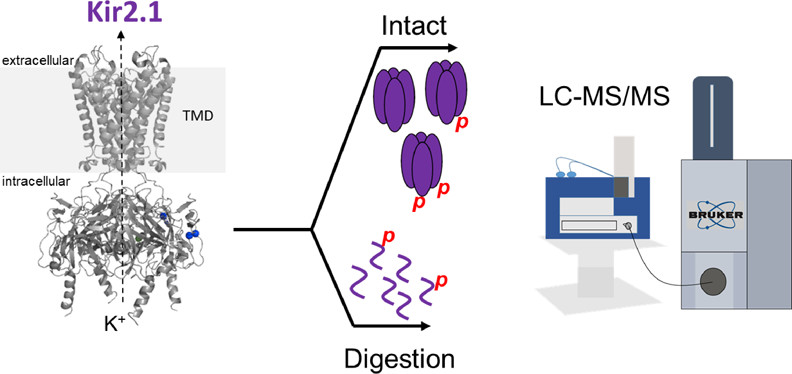
- Brown, K.A.; Anderson, C.; Reilly, L.; Sondhi, K.; Ge, Y.; Eckhardt, L.L. Proteomic Analysis of the Functional Inward Rectifier Potassium Channel (Kir) 2.1 Reveals Several Novel Phosphorylation Sites Biochemistry, 2021, Online ahead of print. Article.
- Bayne, E.B.; Simmons, A.D.; Roberts, D.S.; Zhu, Y.; Aballo, T.A.; Wancewicz, B.; Palecek, S.P.; Ge, Y. Multiomics Method Enabled by Sequential Metabolomics and Proteomics for Human Pluripotent Stem-Cell Derived Cardiomyocytes J. Proteome Res., 2021, 20, 10, 4646-4654. Article.
- Truchan, N.A.; Fenske, R.J.; Sandhu, H.K.; Weeks, A.M.; Patibandla, C.; Wancewicz, B.; Pabich, S.' Reuter, A.; Reuter, A.; Harrington, J.M.; Brill, A.L.; Peter, D.C.; Nall, R.; Daniels, M.; Punt, M.; Kaiser, C.E.; Cox, E.D.; Ge, Y.; Davis, D.B.; Kimple, M.E. Human Islet Expression Levels of Prostaglandin E2 Synthetic Enzymes, But Not Prostaglandin EP3 Receptor, Are Positively Correlated with Markers of β-Cell Function and Mass in Nondiabetic Obesity ACS Pharmacol. Transl. Sci., 2021, 4, 4, 1338-1348. Article.
- Larson, E. J.; Roberts, D. S.; Melby, J. A.; Zhu, Y.; Buck, K. M.; Zhou, S.; Han, L.; Zhang, Q.; Ge, Y. High-Throughput Multi-attribute Analysis of Antibody-Drug Conjugates Enabled by Trapped Ion Mobility Spectrometry and Top-Down Mass Spectrometry Anal. Chem., 2021, 93, 29, 10013-10021. Article.
- Aballo, T. J.; Roberts, D. S.; Melby, J. A.; Buck, K. M.; Brown, K. A; Ge, Y. Ultrafast and Reproducible Proteomics from Small Amounts of Heart Tissue Enabled by Azo and timsTOF Pro J. Proteome Res., 2021, 20, 8, 4203-4211. Article.
- Melby, J.A.; Roberts, D.S.; Larson, E.J.; Brown, K.A.; Bayne, E.B.; Jin, S.; Ge, Y. Novel Strategies to Address the Challenges in Top-Down Proteomics J. Am. Soc. Mass Spectrom., 2021, 32, 6, 1278-1294. Article.
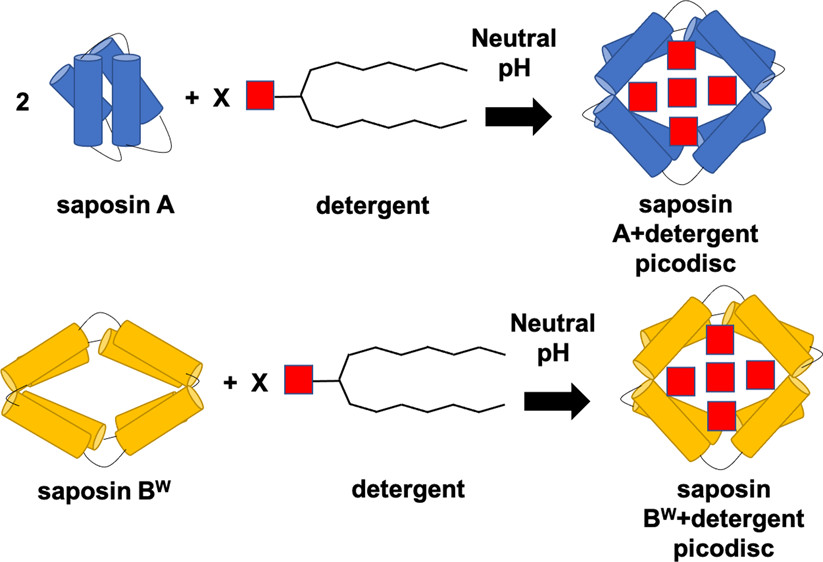
- Kurgan, K.W.; Chen, B.; Brown, K.A.; Falco Cobra, P.; Ye, X.; Ge, Y.; Gellman; S.H. Stable Picodisc Assemblies from Saposin Proteins and Branched Detergents Biochemistry, 2021, 60 (14), 1108-1119. Article.
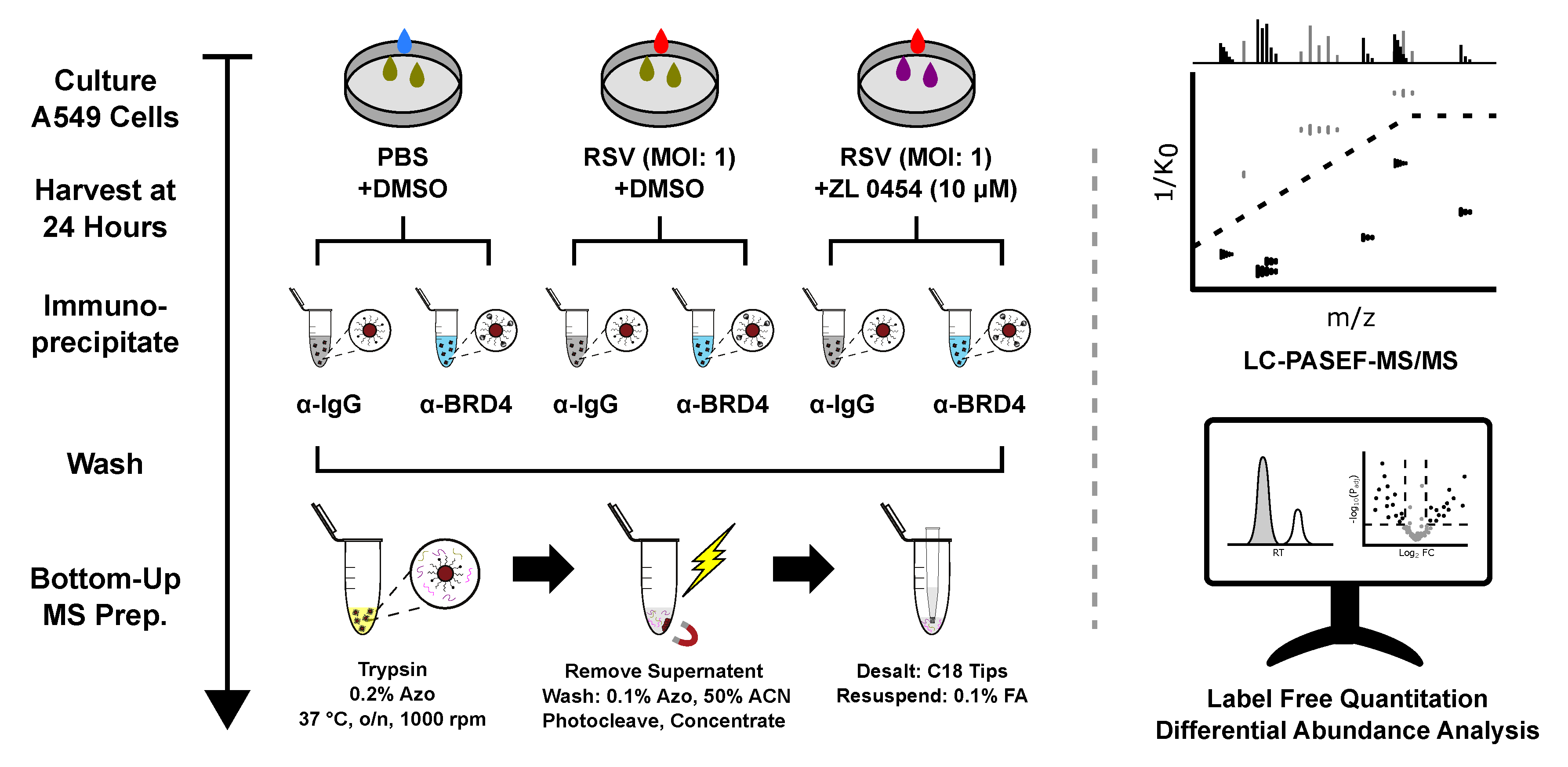
- Mann, M.; Roberts, D.S.; Zhu, Y.; Li, Y.; Zhou, J.; Ge, Y.; Brasier, A.R. Discovery of RSV-Induced BRD4 Protein Interactions Using Native Immunoprecipitation and Parallel Accumulation—Serial Fragmentation (PASEF) Mass Spectrometry Viruses, 2021, 13 (3), 454. Article.
- Melby, J.A.; de Lange, W.J.; Zhang, J.; Roberts, D.S.; Mitchell, S.D.; Tucholski, T.; Kim, G.; Kyrvasilis, A.; McIlwain, S.J.; Kamp, T.J.; Ralphe, J.C.; Ge, Y. Functionally Integrated Top-Down Proteomics for Standardized Assessment of Human Induced Pluripotent Stem Cell-Derived Engineered Cardiac Tissues J. Proteome Res., 2021, 20 (2), 1424-1433. Article.
- Schaid, M.D.; Zhu, Y.; Richardson, N.E.; Patibandla, C.; Ong, I.M.; Fenske, R.J.; Neuman, J.C.; Guthery, E.; Reuter, A.; Sandhu, H.K.; Fuller, M.H.; Cox, E.D.; Davis, D.B.; Layden, B.T.; Brasier, A.R., Lamming, D.W.; Ge, Y.; Kimple, M.E. Systemic Metabolic Alterations Correlate with Islet-Level Prostaglandin E2 Production and Signaling Mechanisms That Predict β-Cell Dysfunction in a Mouse Model of Type 2 Diabetes Metabolites, 2021, 11(1), 58. Article.
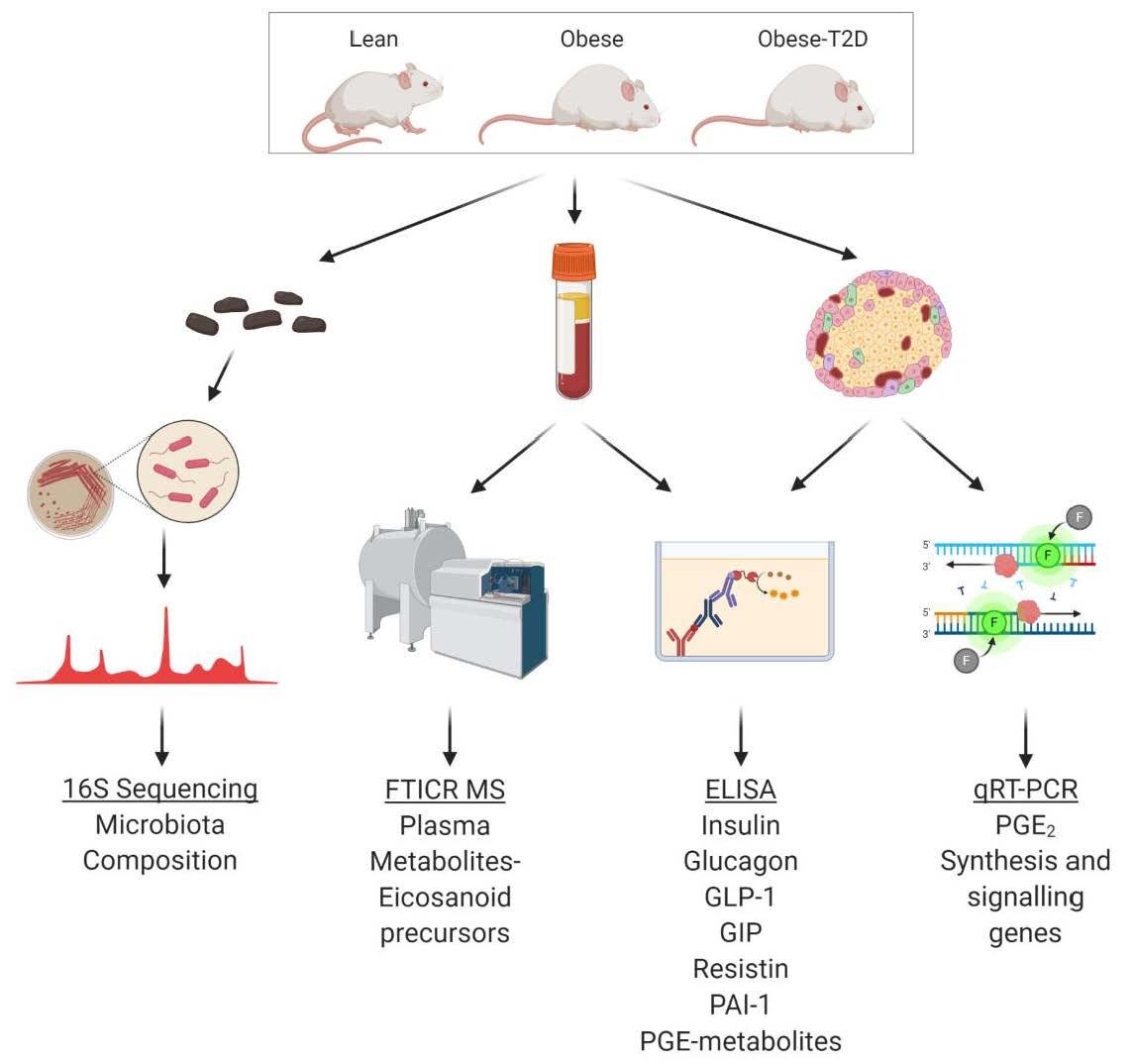
- Brown, K.A.; Melby, J.A.; Roberts, D.S.; Ge, Y. Top-down Proteomics: Challenges, Innovations, and Applications in Basic and Clinical Research Expert Rev Proteomics, 2020, 1-15. Article.
- Knott, S.J.; Brown, K.A.; Josyer, H.; Carr, A.; Inman, D.;Friedl, A.; Ponik, S.M.; Ge, Y. Photocleavable Surfactant-Enabled Extracellular Matrix Proteomics Anal. Chem, 2020, 92 (24), 15693-15698. Article.
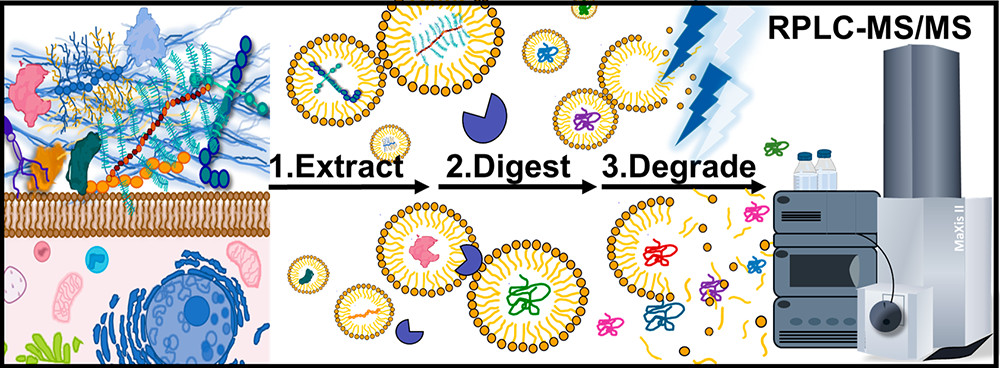
- Brown, K.A.; Tucholski, T.; Alpert, A.J.; Eken, C.; Wesemann, L.; Kyrvalis, A.; Jin, S.; Ge, Y. Top-Down Proteomics of Endogenous Membrane Proteins Enabled by Cloud Point Enrichment and Multidimensional Liquid Chromatography-Mass Spectrometry Anal. Chem, 2020, 92 (24), 15726-15735. Article.
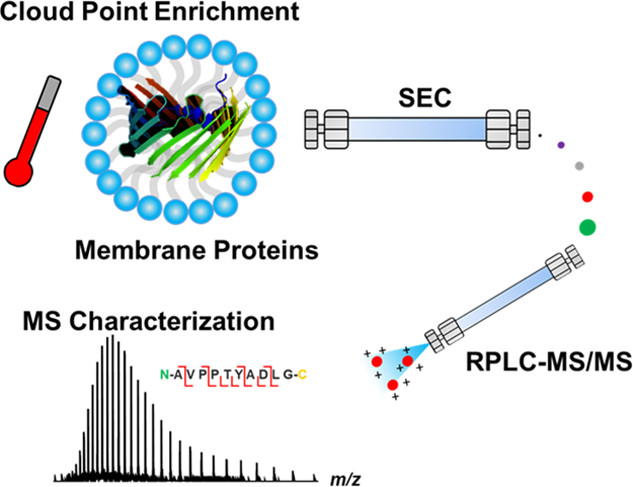
- Styles, M.J.; Early, S.A.; Tucholski, T.; West, K.H.J.; Ge, Y.; Blackwell, H.E. Chemical Control of Quorum Sensing in E. coli: Identification of Small Molecule Modulators of SdiA and Mechanistic Characterization of a Covalent Inhibitor, ACS Infect. Dis., 2020, 6 (12), 3092-3103. Article.
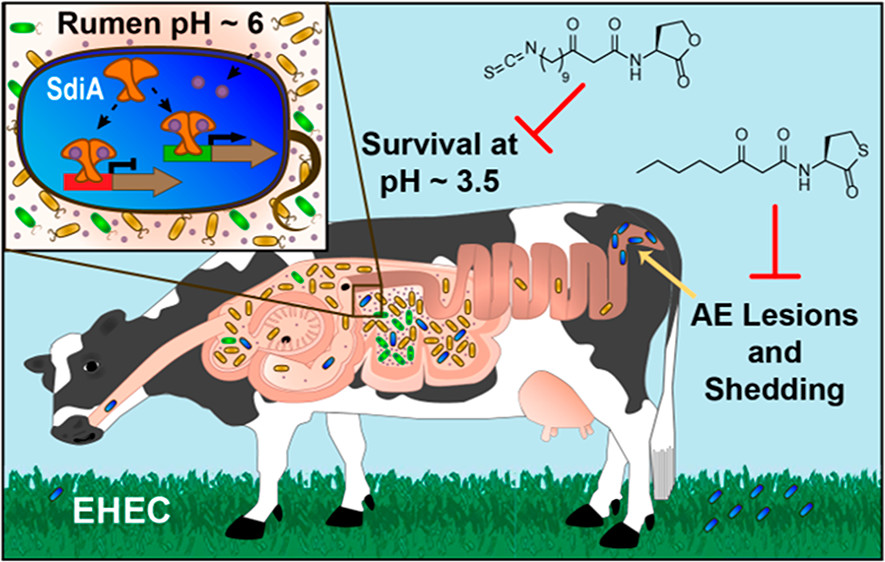
- Larson, E.J.; Zhu, Y.; Wu, Z.; Chen, B.; Zhang, Z.; Zhou, S.; Han, L.; Zhang, Q.; Ge, Y.; Rapid Analysis of Reduced Antibody Drug Conjugate by Online LC-MS/MS with Fourier Transform Ion Cyclotron Resonance Mass Spectrometry, Anal. Chem., 2020, 92, 22, 15096-15103. Article.
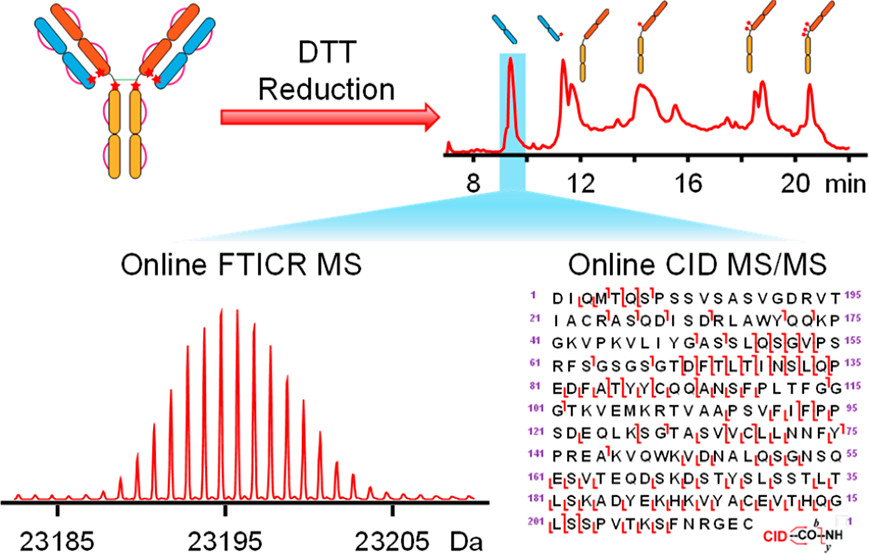
- Zhou, M.; Lantz, C.; Brown, K.A.; Ge, Y.; Pasa-Tolic, L.; Loo, J.A.; Lermyte, F.; Higher-order structural characterisation of native proteins and complexes by top-down mass spectrometry, Chem. Science, 2020, 11, 12918-12936. Article.
- Zhu, Y.; Wancewicz, B.; Schaid, M.; Tiambeng, T.N.; Wenger, K.; Jin, Y.; Heyman, H.; Thompson, C.J.; Barsch, A.; Cox, E.D.; Davis, D.B.; Brasier, A.R.; Kimple, M.; Ge, Y. Ultrahigh-Resolution Mass Spectrometry-Based Platform for Plasma Metabolomics Applied to Type 2 Diabetes Research, J. Proteome Res., 2020, 20 (1), 463-473. Article.
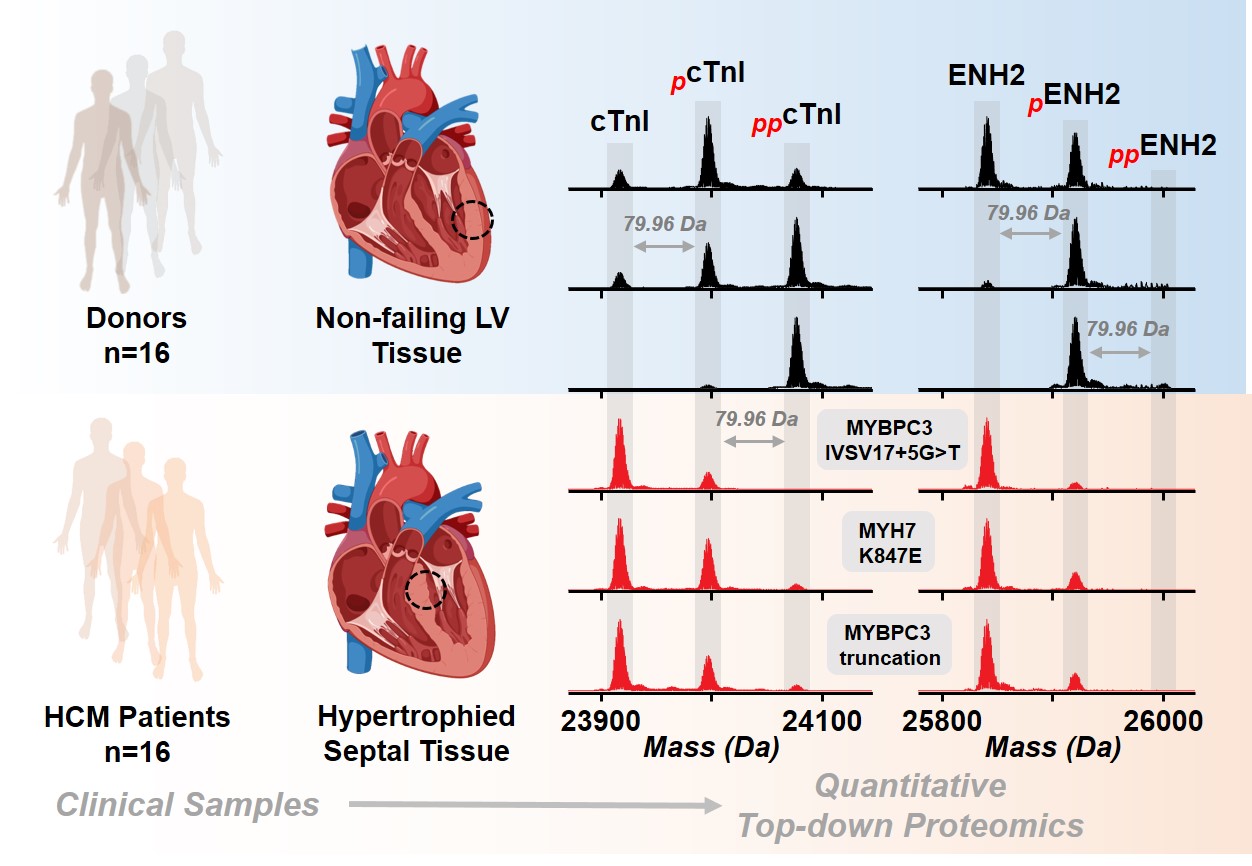
- Tucholski, T.; Cai, W.; Gregorich, Z.R.; Bayne, E.F.; Mitchell, S.D.; McIlwain, S.J.; de Lange, W.J.; Wrobbel, M.; Karp, H.; Hite, Z.; Vikhorev, P.G.; Marston, S.B.; Lal, S.; Li, A.; dos Remedios, C.; Kohmoto, T.; Hermsen, J.; Ralphe, J.C.; Kamp, T.J.; Moss, R.L.; Ge, Y. Distinct hypertrophic cardiomyopathy genotypes result in convergent sarcomeric proteoform profiles revealed by top-down proteomics, Proc Natl Acad Sci USA, 2020, 117 (40), 24691-24700. Article.
- Kellie, J.F.; Tran, J.C.; Jian, W.; Jones, B.; Mehl, J.T.; Ge, Y.; Henion, J.; Bateman, K.P. Intact Protein Mass Spectrometry for Therapeutic Protein Quantitation, Pharmacokinetics, and Biotransformation in Preclinical and Clinical Studies: An Industry Perspective, J. Am. Soc. Mass Spectrom., 2020, 32 (8), 1886-1900. Article.
- Tucholski, T.; Ge, Y. Fourier‐transform ion cyclotron resonance mass spectrometry for characterizing proteoforms, Mass Spectrom. Rev., 2020, 41 (2), 158-177. Article.
- Tiambeng, T. N.*; Roberts, D. S.*; Brown, K.A.; Zhu, Y.; Chen, B.; Wu, Z.; Mitchell, S. D.; Guardado-Alvarez, T.M.; Jin, Y.; Ge, Y. Nanoproteomics enables proteoform-resolved analysis of low-abundance proteins in human serum., Nat Commun., 2020, 11 (1), 1-12. Article.
- Wu, Z.; Roberts, D.S.; Melby, J.A.; Wenger, K.; Wetzel, M.; Gu, Y.; Ramanathan, S.G.; Bayne, E.F.; Liu, X.; Sun, R.; Ong, I.M.; McIlwain, S.J.; Ge, Y. MASH Explorer: A Universal Software Environment for Top-Down Proteomics., J. Proteome Res., 2020, 19, 9, 3867-3876. Article.

- Srzentic, K.; Fornelli, L.; Tsybin, Y.O.; Loo, J.A.; Seckler, H.; Agar, J.N.; Anderson, L.C.; Bai, D.L.; Beck, A.; Brodbelt, J.S.; van der Burgt, Y.E.M.; Chamot-Rooke, J.; Chatterjee, S.; Chen, Y.; Clarke, D.J.; Danis, P.O.; Diedrich, J.K.; D'Ippolito, R.A.; Dupré, M.; Gasilova, N.; Ge, Y.; Ah Goo, Y.; Goodlett, D.R.; Greer, S.M.; Haselmann, K.F.; He, L.; Hendrickson, C.L.; Hinkle, J.D.; Holt, M.; Hughes, S.; Hunt, D.F.; Kelleher, N.L.; Kozhinov, A.N.; Lin, Z.; Malosse, C.; Marshall, A.G.; Menin, L.; Millikin, R.J.; Nagornov, K.O.; Nicolardi, S.; Paša-Tolić, L.; Pengelley, S.; Quebbemann, N.R.; Resemann, A.; Sandoval, W.; Sarin, R.; Schmitt, N.D.; Shabanowitz, J.; Shaw, J.B.; Shortreed, M.R.; Smith, L.M.; Sobott, F.; Suckau, D.; Toby, T.; Weisbrod, C.R.; Wildburger, N.C.; Yates, J.R.; Yoon, S.H.; Young, N.L.; Zhou, M. Inter-laboratory Study for Characterizing Monoclonal Antibodies by Top-Down and Middle-Down Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2020, 31, 9, 1783-1802. Article.
- Xu, B.; Li, M.; Wang, Y.; Zhao, M.; Morotti, S.; Shi, Q.; Wang, Q.; Barbagallo, F.; Teoh, J.; Reddy, G.R.; Bayne, E.F.; Liu, Y.; Shen, A.; Puglisi, J.L. Ge, Y.; Li, J.; Grandi, E.; Nieves-Cintron, M.; Xiang, Y.K. GRK5 Controls SAP97-Dependent Cardiotoxic β1 Adrenergic Receptor-CaMKII Signaling in Heart Failureβ, Circ Res., 2020, 127, 796-810. Article.
- McIlwain, S.; Wu, Z.; Wetzel, M.; Belongia, D.; Jin, Y.; Wenger, K.; Ong, I.; Ge, Y. Enhancing Top-Down Proteomics Data Analysis by Combining Deconvolution Results through a Machine Learning Strategy., J. Am. Soc. Mass Spectrom., 2020, 31(5), 1104-1113. Article.
- Brown, K; Tucholski, T; Eken, C; Knott, S; Zhu, Y; Jin, S; Ge, Y. High-throughput Proteomics Enabled by a Photocleavable Surfactant., Angew Chem Int Ed Engl., 2020, 59(22), 8406-8410. Article.
- Zhang, F; Wyche, TP; Zhu, Y; Braun, DR; Yan, JX; Chanana, S; Ge, Y.; Guzei, IA; Chevrette, MG; Currie, CR; Thomas, MG; Rajski, SR; Bugni, TS.; MS-Derived Isotopic Fine Structure Reveals Forazoline A as a Thioketone-Containing Marine-Derived Natural Product., Org Lett., 2020, 22(4), 1275-1279. Article.
- Melby, J. A.; Jin, Y.; Lin, Z.; Tucholski, T.; Wu, Z.; Gregorich, Z. R.; Diffee, G. M.; Ge, Y. Top-Down Proteomics Reveals Myofilament Proteoform Heterogeneity among Various Rat Skeletal Muscle Tissues., J. Proteome Res. 2020, 19(1), 443-454. (Article selected for cover of Jan. issue) Article.

- Bortnov, V.; Tonelli, M.; Lee, W.; Lin, Z.; Annis, D. S.; Demerdash, O.; Bateman, A.; Mitchell, J.; Ge, Y.; Markley, J. L.; Mosher, D. F. Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum, Nat Commun., 2019, 10, 1-14. Article.
- Cai, W.; Zhang, J.; De Lange, W. J.; Gregorich, Z. R.; Karp, H.; Ferrell, E. T.; Mitchell, S. T.; Tucholski, T.; Lin, Z.; Biermann, M.; McIlwain, S.; Ralphe, J. C.; Kamp, T. J.; Ge, Y. Unbiased Proteomics Method to Assess the Maturation of Human Pluripotent Stem Cell-Derived Cardiomyocytes., Circ Res., 2019, 125, 936-953. Article.
- Tiambeng, T. N.; Tucholski, T.; Wu, Z.; Zhu, Y.; Mitchell, S. D.; Roberts, D. S.; Jin, Y.; Ge, Y. Analysis of cardiac troponin proteoforms by top-down mass spectrometry, Methods Enzymol., ISSN 0076-6879 Article.
- Wu, Z.; Jin, Y.; Chen, B.; Gugger, M. K.; Wilkinson-Johnson, C. L.; Tiambeng, T. N.; Jin, S.; Ge, Y., Comprehensive Characterization of Recombinant Catalytic Subunit of cAMP-Dependent Protein Kinase by Top-Down Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2019, 30, 2561-2570. Article.
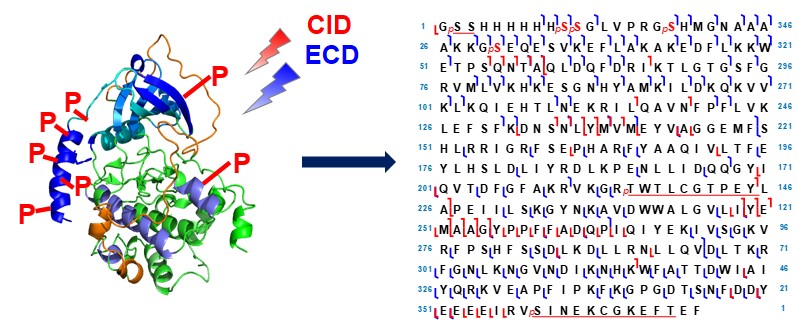
- Smith, L. M.; Thomas, P. M.; Shortreed, M. R.; Scheffer, L. V.; Fellers, R. T.; LuDec, R. D.; Tucholski, T.; Ge, Y.; Agar, J. N.; Anderson, L. C.; Chamot-Rooke, J.; Gault, J.; Loo, J. A.; Pasa-Tolic, L.; Robinson, C. V.; Schluter, H.; Tsybin, Y. O.; Vilaseca, M.; Vizcaino, J. A.; Danis, P. O.; Kelleher, N. L., A five-level classification system for proteoform identifications, Nature Methods. 2019, 16, 939-940. Article.
- Chen, B.; Lin, Z.; Zhu, Y.; Jin, Y.; Larson, E.; Xu, Q.; Fu, C.; Zhang, Z.; Zhang, Q.; Pritts, W. A.; Ge, Y. Middle-Down Multi-Attribute Analysis of Antibody-Drug Conjugates with Electron Transfer Dissociation., Anal Chem. 2019, 16, 18, 11661-11669. Article.
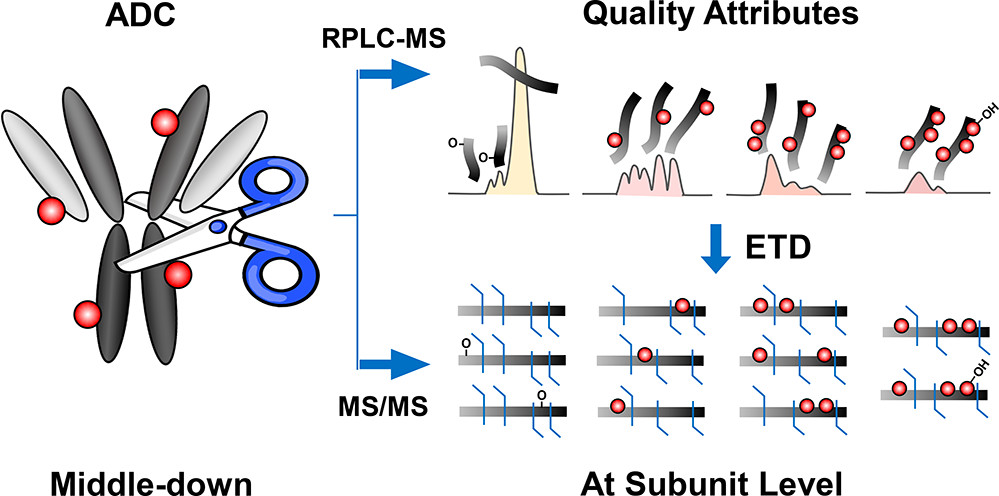
- Schaffer, L. A.; Tucholski, T.; Shortreed, M. R.; Ge, Y.; Smith, L. M., Intact-Mass Analysis Facilitating the Identification of Large Human Heart Proteoforms., Anal Chem. 2019, 19, 10937-10942. Article.
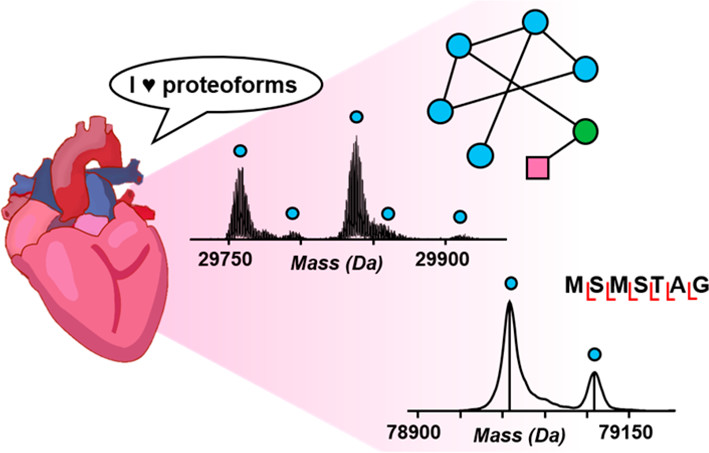
- Biermann, M.; Cai, W.; Lang, D.; Hermsen, J.; Profio, L.; Zhou, Y.; Czirok, A.; Isai, D. G.; Napiwocki, B. N.; Rodriguez, A. M.; Brown, M. E.; Woon, M. T.; Shao, A.; Han, T.; Park, D.; Hacker, T. A.; Crone, W. C.; Burlingham, W. J.; Glukhov, A. V.; Ge, Y.; Kamp, T. J., Epigenetic Priming of Human Pluripotent Stem Cell-Derived Cardiac Progenitor Cells Accelerates Cardiomyocyte Maturation, Stem Cells. 2019, 37, 910-923. Article.
- Brown, K. A.; Chen, B.; Guardado-Alvarez, T. M.; Lin, Z.; Hwang, L.; Ayaz-Guner, S.; Jin, S.; Ge, Y. A Photocleavable Surfactant for Top-Down Proteomics, Nature Methods. 2019, 16, 417-420. Article. News Release.
- Donnelly, D. P.; Rawlins, C. M.; DeHart, C. J.; Fornelli,, L.; Schachner, L. F.; Lin, Z.; Lippens, J. L.; Aluri, K. C.; Sarin, R.; Chen, B.; Lantz, C.; Jung, W.; Johnson, K. R.; Koller, A.; Wolff, J. J.; Campuzano, I. D. G.; Auclair, J. R.; Ivanov, A. R.; Whitelegge, J. P.; Paša-Tolić, L.; Chamot-Rooke, J.; Danis, P. O.; Smith, L. M.; Tsybin, Y. O.; Loo, J. A.; Ge Y.; Kelleher N. L.; Agar J. N. Best practices and benchmarks for intact protein analysis for top-down mass spectrometry., Nature Methods. 2019, 16, 587-594. Article.
- Schaffer, L. V.; Millikin, R. J.; Miller, R. M.; Anderson, L. C.; Fellers R. T.; Ge Y.; Kelleher N. L.; LeDuc R. D.; Liu X.; Payne, S. H.; Sun, L.; Thomas, P. M.; Tucholski, T.; Wang, Z.; Wu, S.; Wu, Z.; Yu, D.; Shortreed, M. R.; Smith, L. M. Identification and Quantification of Proteoforms by Mass Spectrometry, Proteomics, 2019, 19, 1800361 Article.
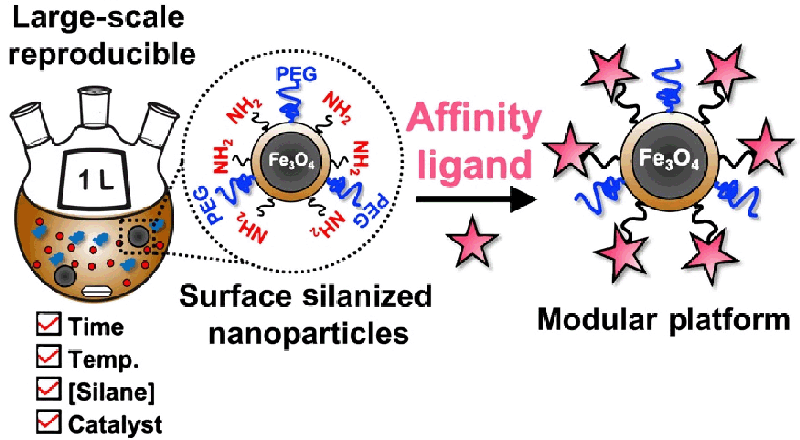
- Roberts, D. S.; Chen, B.; Tiambeng, T. N.; Wu, Z.; Ge, Y.; Jin, S. Reproducible large-scale synthesis of surface silanized nanoparticles as an enabling nanoproteomics platform: Enrichment of the human heart phosphoproteome, Nano Res., 2019, 12, 1473-1481. DOI: 10.1007/s12274-019-2418-4. Article.
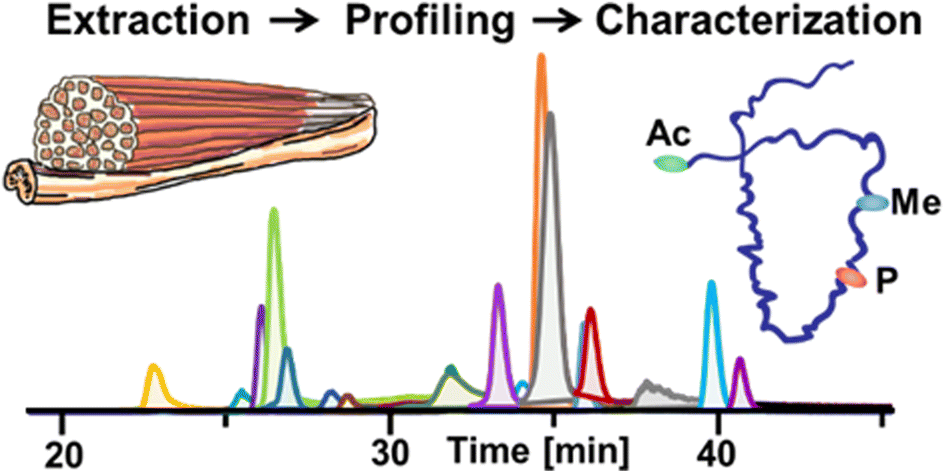
- Jin, Y.; Diffee, G. M.; Colman, R. J.; Anderson, R. M.; Ge, Y. Top-down Mass Spectrometry of Sarcomeric Protein Post-translational Modifications from Non-human Primate Skeletal Muscle, J. Am. Soc. Mass Spectrom. 2019. Epub ahead of print. DOI: 10.1007/s13361-019-02139-0 Article
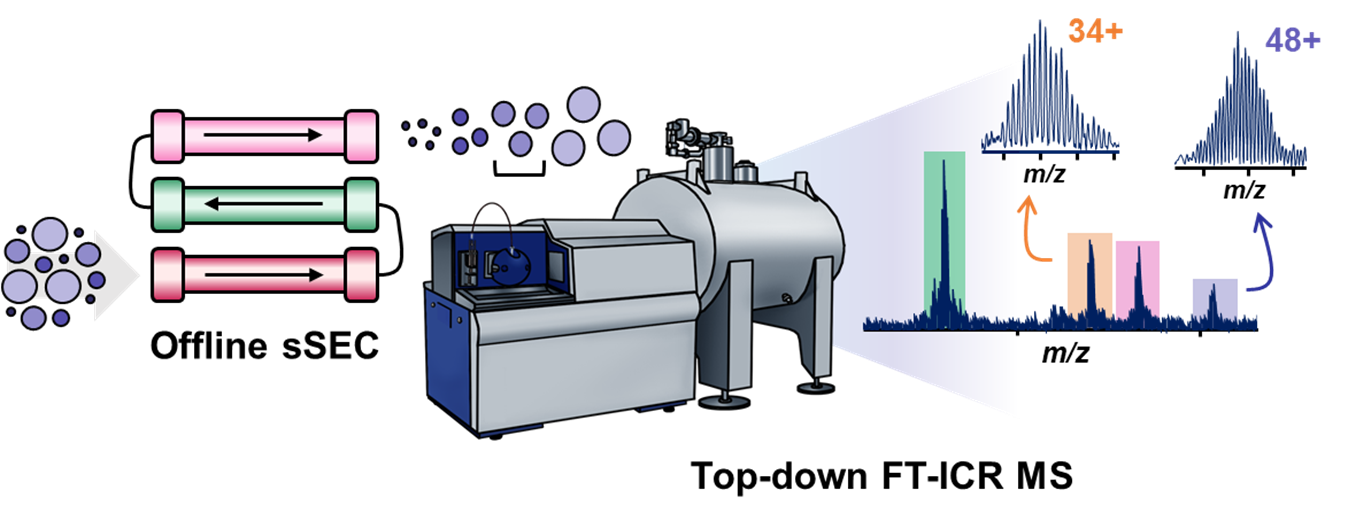
- Tucholski, T.; Knott, S.; Chen, B.; Pistono, P.; Lin, Z.; Ge, Y. A Top-Down Proteomics Platform Coupling Serial Size Exclusion Chromatography and Fourier Transform Ion Cyclotron Resonance Mass Spectrometry, Anal Chem., 2019, 91, 3855-3844. DOI: 10.1021/acs.analchem.8b04082. Article
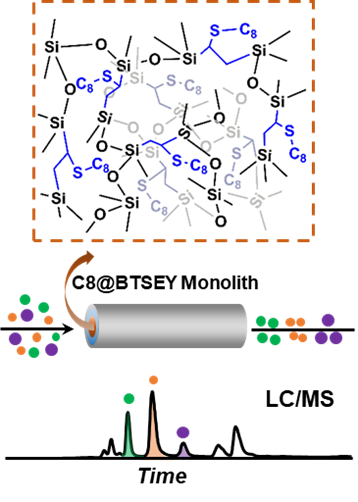
- Liang, Y.; Jin, Y.; Wu, Z.; Tucholski, T.; Brown, K.A.; Zhang, L.; Zhang, Y.; Ge, Y. Bridged Hybrid Monolithic Column Coupled to High-Resolution Mass Spectrometry for Top-down Proteomics, Anal Chem., 2019, 91, 3, 1743-1747. DOI: 10.1021/acs.analchem.8b05817. Article
- Gregorich, Z.R.; Patel, J.R.; Cai, W.; Lin, Z.; Heurer, R.; Fitzsimons, D.P.; Moss, R.L.; Ge, Y. Deletion of Enigma Homologue from the Z-disc slows tension development kinetics in mouse myocardium, J Gen Physiol., 2019. PII: jgp.201812214. DOI: 10.1085/jgp.201812214. Article
- Berry, J.L.; Zhu, W.; Tang, Y.L.; Krishnamurthy, P.; Ge, Y.; Cooke, J.P.; Chen, Y.; Garry, D.J.; Yang, H.T.; Rajasekaran, N.S.; Koch, W.J.; Li, S., Domae, K.; Qin, G.; Cheng, K.; Kamp, T.J.; Ye, L.; Hu, S.; Ogle, B.M.; Rogers, J.M.; Abel, E.D.; Davis, M.E.; Prabhu, S.D.,; Liao, R.; Pu, W.T.; Wang, Y.; Ping, P.; Bursac, N.; Vunjak-Novakovic, G.; Wu, J.C.; Bolli, R.; Menasché, P.; Zhang, J. Convergences of Life Sciences and Engineering in Understanding and Treating Heart Failure, Circ Res., 2019, 124, 1, 161-169. DOI: 10.1161/CIRCRESAHA.118.314216. Article
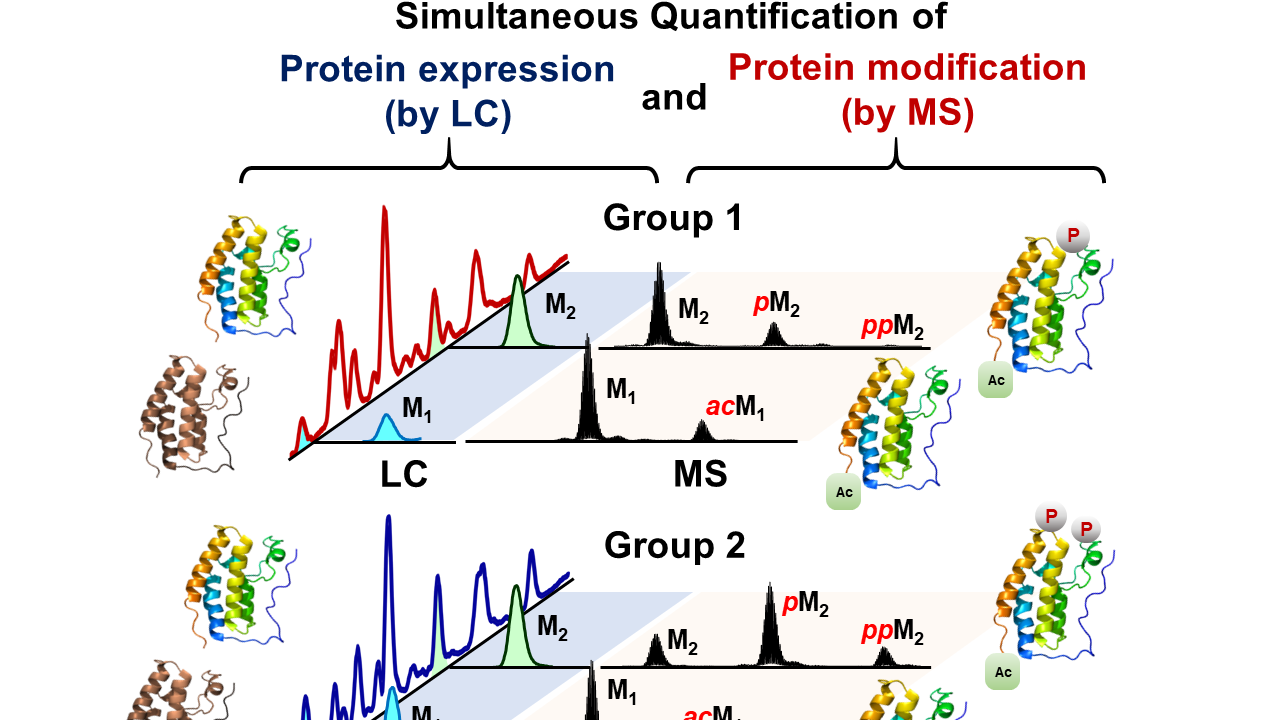
- Lin, Z.; Wei, L.; Cai, W.; Zhu, Y.; Tucholski, T.; Mitchell, S.D.; Guo, W.; Ford, S.P.; Diffee, G.M.; Ge, Y.
Simultaneous Quantification of Protein Expression and Modifications by Top-down Targeted Proteomics: A Case of Sarcomeric Subproteome, Mol Cell Proteomics., 2019, 18, 3, 594-605. PII: mcp.TIR118.001086. DOI: 10.1074/mcp.TIR118.00108.
Article
- Jin, Y.; Lin, Z.; Xu, Q.; Fu, C.; Zhang, Z.; Zhang, Q.; Pritts, W.A.; Ge, Y. Comprehensive characterization of monoclonal antibody by Fourier transform ion cyclotron resonance mass spectrometry, MAbs., 2019, 11, 1, 106-115. DOI: 10.1080/19420862.2018.1525253. Article
- Chen, Z.; Maimaiti, R.; Zhu, C.; Cai, H.; Stern, A.; Mozdziak, P.; Ge, Y.; Ford, S.P.; Nathanielsz P.W.; Guo, W.; Z- band and M-band titin splicing and regulation by RNA binding motif 20 in striated muscles, J. Cell Biochem., 2018, 119, 12, 9986-9996. DOI: 10.1002/jcb.27328. Article
- Cai, W.; Hite, Z.L.; Lyu, B.; Wu, Z.; Lin, Z.; Gregorich, Z. R.; Messer, A.E.; McIlwain, S.J.; Marston, S.B.; Kohmoto, T.; Ge, Y.
Temperature-sensitive sarcomeric protein post-translational modifications revealed by top-down proteomics, J. Mol. Cell Cardiol., 2018, 122, 11-22. DOI: 10.1016/j.yjmcc.2018.07.247
Article
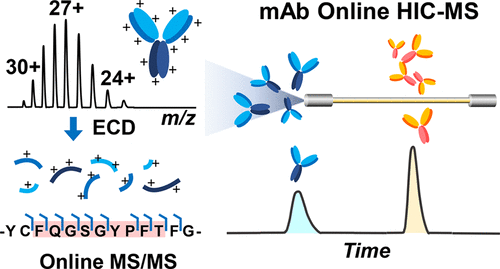
- Chen, B.; Lin, Z.; Alpert, A. J.; Fu, C.; Zhang, Q.; Pritts, W. A.; Ge, Y.
Online hydrophobic interaction chromatography-mass spectrometry for the analysis of intact monoclonal antibodies, Anal. Chem., 2018 , 90, 12, 7135-7138 DOI:10.1021/acs.analchem.8b01865
Article
- Martin-Garrido, A.; Biesiadecki, B.J.; Salhi, H.E.; Shaifta, Y.; dos Remedios, C.G.; Ayuz-Guner, S.; Cai, W.; Ge, Y.; Avkiran, M.; Kentish, J.C., Monophosphorylation of cardiac troponin-I at Ser23/24 is sufficient to regulate cardiac myofibrillar Ca2+ sensitivity and calpain-induced proteolysis, J. Biol. Chem., 2018 , 293, 22, 8588-8599, DOI:10.1074/jbc.RA117.001292 Article
- Wu, Z.; Tiambeng, T. N.; Cai, W.; Chen, B.; Lin, Z.; Gregorich, Z. R.; Ge, Y.,
Impact of phosphorylation on the mass spectrometry quantification of intact phosphoproteins, Anal. Chem., 2018, 90, 8, 4935-4939, DOI:10.1021/acs.analchem.7b05246
Article
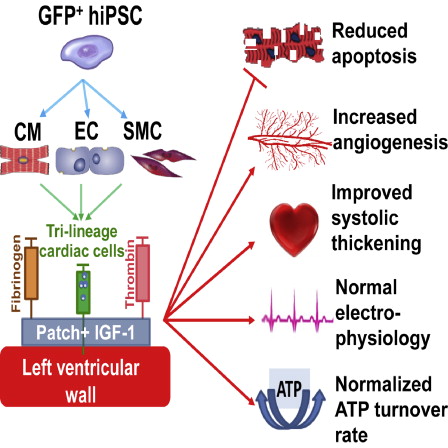
- Gao, L.; Gregorich, Z. R.; Zhu, W.; Mattapally, S.; Lou, X.; Borovjagin, A. V.; Walcott, G. P.; Pollard, A. E.; Fast, V. G.; Ge, Y.; Zhang, J.
Large cardiac-muscle patches engineered from human induced-pluripotent stem-cell-derived cardiac cells improve recovery from myocardial infarction in swine, Circulation, 2018, 137, 16, 1712-1730, DOI:10.1161/CIRCULATIONAHA.117.030785
Article

- Smelter, D. F., De Lange, W. J., Cai, W., Ge, Y., Ralphe, J. C., The HCM-linked W792R mutation in cardiac myosin binding protein-C reduces C6 FnIII domain stability. Am J Physiol Heart Circ Physiol. 2018, 314, 6, H1179-H1191. DOI: 10.1152/ajpheart.00686.2017. Article
- Lin, Z.; Guo, F.; Gregorich, Z. R.; Sun, R.; Zhang, H.; Hu, Y.; Shanmuganayagam, D.; Ge, Y., Comprehensive characterization of swine cardiac troponin T proteoforms by top-down mass spectrometry, J. Am. Soc. Mass Spectrom. 2018, Accepted.
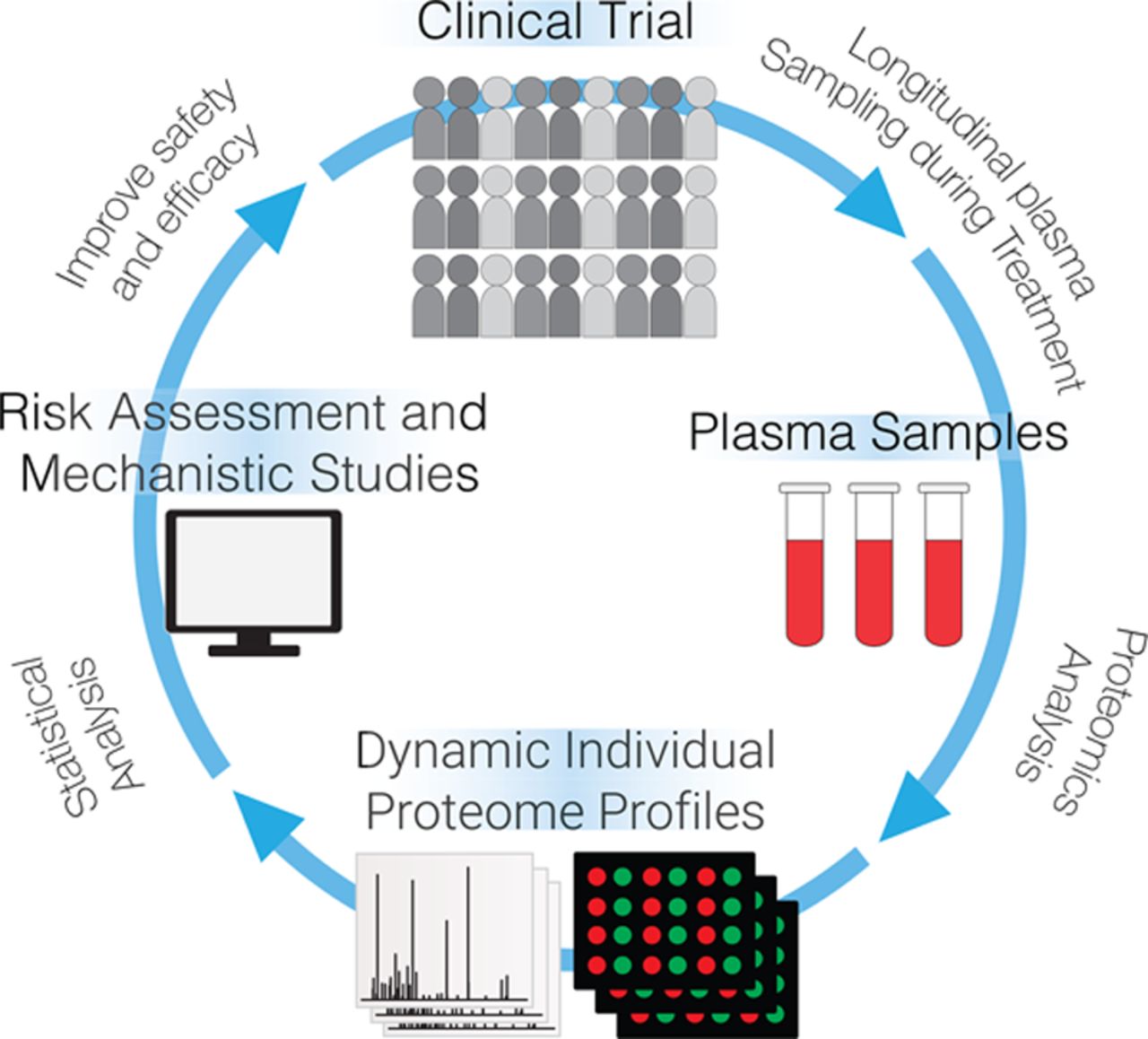
- Lam, M. P. Y.; Ge, Y., Harnessing the power of proteomics to assess drug safety and guide clinical trials, Circulation, 2018, 137, 1011-1014. Article
- Aebersold, R.; Agar, J. N.; Amster, I. J.; Baker, M. S.; Bertozzi, C. R.; Boja, E. S.; Costello, C. E.; Cravatt, B. F.; Fenselau, C.; Garcia, B. A.; Ge, Y.; Gunawardena, J.; Hendrickson, R. C.; Hergenrother, P. J.; Huber, C. G.; Ivanov, A. R.; Jensen, O. N.; Jewett, M. C.; Kelleher, N. L.; Kiessling, L. L.; Krogan, N. J.; Larsen, M. R.; Loo, J. A.; Ogorzalek Loo, R. R.; Lundberg, E.; MacCoss, M. J.; Mallick, P.; Mootha, V. K.; Mrksich, M.; Muir, T. W.; Patrie, S. M.; Pesavento, J. J.; Pitteri, S. J.; Rodriguez, H.; Saghatelian, A.; Sandoval, W.; Schlüter, H.; Sechi, S.; Slavoff, S. A.; Smith, L. M.; Snyder, M. P.; Thomas, P. M.; Uhlén, M.; Van Eyk, J. E.; Vidal, M.; Walt, D. R.; White, F. M.; Williams, E. R.; Wohlschlager, T.; Wysocki, V. H.; Yates, N. A.; Young, N. L.; Zhang, B., How many human proteoforms are there? Nat. Chem. Biol. 2018, 14, 206-214. doi: 10.1038/nchembio.2576. Article
- LeDuc, R.D.; Schwämmle, V.; Shortreed, M.R.; Cesnik, A.J.; Solntsev, S.K.; Shaw JB, Martin MJ, Vizcaino JA, Alpi E, Danis P, Kelleher NL, Smith LM, Ge, Y., Agar JN, Chamot-Rooke J, Loo JA, Pasa-Tolic L, Tsybin YO. ProForma: A Standard Proteoform Notation. J. Proteome Res. 2018, 17, 1321-1325. doi: 10.1021/acs.jproteome.7b00851. Article

- Chen, Z., Song, J., Chen, L., Zhu, C., Cai, H., Sun, M., Stern, A., Mozdziak, P., Ge, Y., Means, W. J., Guo, W., Characterization of TTN Novex Splicing Variants across Species and the Role of RBM20 in Novex-Specific Exon Splicing. Genes, 2018, 9, 86. DOI: 10.3390/genes9020086 Article
- Seok, S.H.; Ma, Z.X.; Feltenberger, J. B.; Chen, H.; Chen, H.; Scarlett, C.; Lin, Z.; Satyshur, K.A.; Cortopassi, M.; Jefcoate, C.R.; Ge, Y.; Tang, W.; Bradfield, C. A.; Xing, Y. Trace derivatives of kynurenine potently activate the aryl hydrocarbon receptor (AHR). J Biol Chem. 2018, 293, 1994-2005. DOI: 10.1074/jbc.RA117.000631 Article
- Chen, B.; Brown, K.A.; Lin, Z.; Ge, Y.
Top-down proteomics: ready for prime time?
Anal. Chem., 2018, 90, 1, 110-127, DOI: 10.1021/acs.analchem.7b04747.
Article
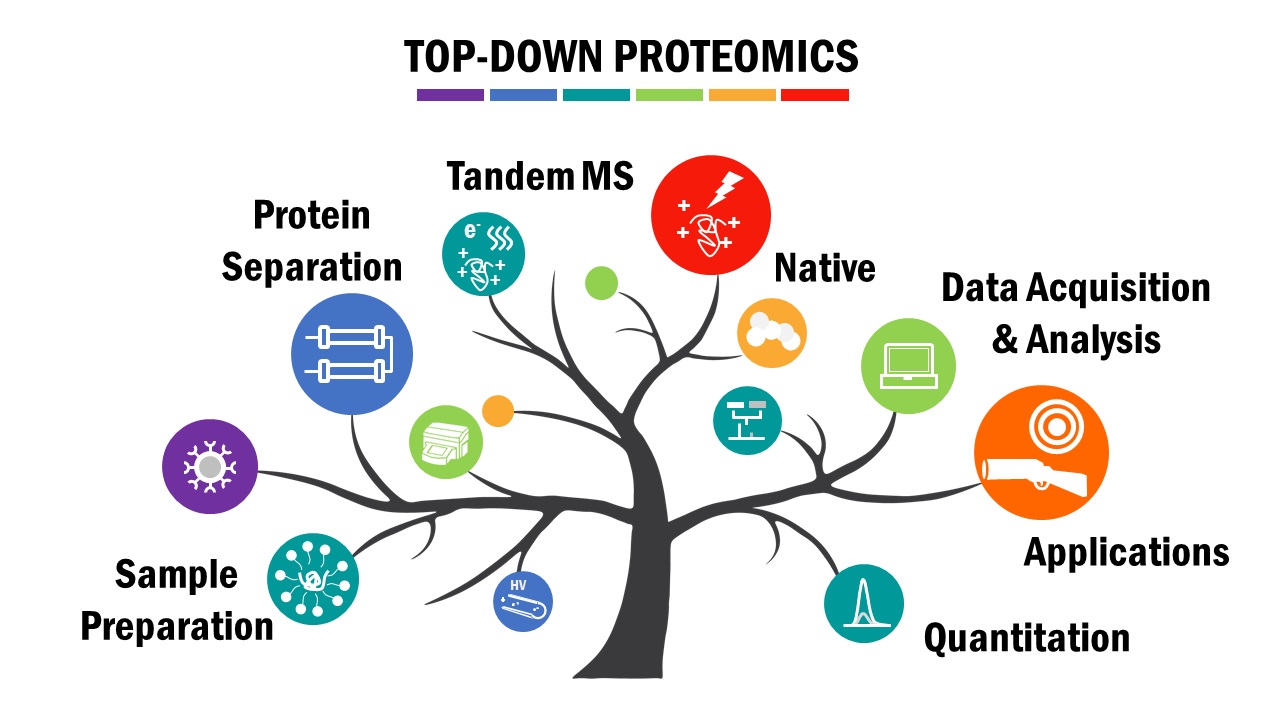
- Wei, L.; Gregorich, Z. R.; Lin, Z.; Cai, W.; Jin, Y.; McKiernan, S. H.; Mcilwain, S.; Aiken, J. M.; Moss, R. L.; Diffee, G. M.; Ge, Y.
Novel sarcopenia-related alterations in sarcomeric protein post-translational modifications in skeletal muscles identified by top-down proteomics,
Mol. Cell. Proteomics, 2018, 17, 134-145, DOI: 10.1074/mcp.RA117.000124
Article

- Hu, C.; Worth, M.; Fan, D.; Li, B.; Li, H.; Lu, L. Zhong, X.; Lin, Z.; Wei, L.; Ge, Y.; Li, L.; Jiang, J. Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase, Nat. Chem. Biol., 2017, 13, 1267-1273. DOI: 10.1038/nchembio.2494. Article
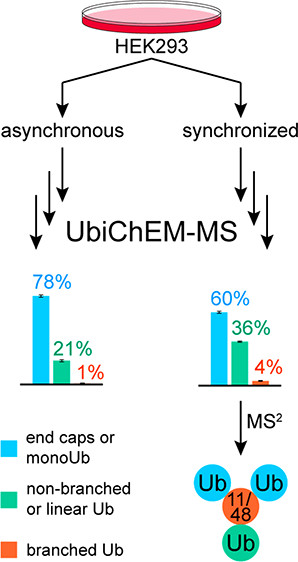
- Rana, A. S. J. B.; Ge, Y.; Strieter, E. R.
Ubiquitin chain enrichment middle-down mass spectrometry (ubichem-ms) reveals cell-cycle dependent formation of lys11/lys48 branched ubiquitin chains.
J. Proteome Res.. 2017, 16 , 3363-3369. DOI: 10.1021/acs.jproteome.7b00381.
Article
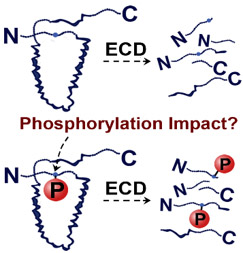
- Chen, B.; Guo, X.; Tucholski, T.; Lin, Z.; Mcilwain, S.; Ge, Y.
The impact of phosphorylation on electron capture dissociation of proteins: a top-down perspective.
J. Am. Soc. Mass Spectrom., 2017, 28, 1805-1814. DOI: 10.1007/s13361-017-1710-3
Article
- Wu, C.-G., Chen, H., Guo, F., Yadav, V. K., McIlwain, S. J., Rowse, M., Choudhary, A., Lin, Z., Li, Y., Gu, T., Zheng, A., Xu, Q., Lee, W., Resch, E., Johnson, B., Day, J., Ge, Y., Ong, I. M., Burkard, M. E., Ivarsson, Y., Xing, Y.
PP2A-B′ holoenzyme substrate recognition, regulation and role in cytokinesis. Cell Disc. 2017, 3, 17027. DOI:10.1038/celldisc.2017.27
Article
-
Gregorich Z. R.; Cai, W.; Lin, Z.; Chen, A. J.; Peng, Y.; Kohmoto, T.; Ge, Y.
Distinct sequences and post-translational modifications in cardiac atrial and ventricular myosin light chains revealed by top-down mass spectrometry.
J. Mol. Cell. Cardiol., 2017, 107, 13-21. DOI: 10.1016/j.yjmcc.2017.04.002.
Article
-
Cai, W.; Tucholski, T.; Chen, B.; Alpert, A. J.; McIlwain, S.; Kohmoto, T.; Jin, S.; Ge, Y.
Top-down Proteomics of Large Proteins up to 223 kDa Enabled by Serial Size Exclusion Chromatography Strategy.
Anal Chem., 2017, 89, 5467-5475. DOI: 10.1021/acs.analchem.7b00380.
Article
-
Jin, Y.; Wei, L.; Cai, W.; Lin, Z.; Wu, Z.; Peng, Y.; Kohmoto, T.; Moss, R. M.; Ge, Y.
Complete Characterization of Cardiac Myosin Heavy Chain (223 kDa) Enabled by Size-Exclusion Chromatography and Middle-down Mass Spectrometry.
Anal Chem., 2017, 89, 4922-4930. DOI: 10.1021/acs.analchem.7b00113.
Article
-
Chen, B.; Hwang, L.; Ochowicz, W.; Lin, Z.; Guardado-Alvarez, T. M.; Cai, W.; Xiu, L.; Dani, K.; Colah, C.; Jin, S.; Ge, Y.
Coupling Functionalized Cobalt Ferrite Nanoparticle Enrichment with Online LC/MS/MS for Top-down Phosphoproteomics.
Chem. Sci., 2017, DOI: 10.1039/C6SC05435H.
Article
-
Yang, L; Gregorich Z. R.; Cai, W.; Zhang, P; Young, B; Gu, Y; Zhang, J; Ge, Y.
Quantitative Proteomics and Immunohistochemistry Reveal Insights into Cellular and Molecular Processes in the Infarct Border Zone One Month after Myocardial Infarction.
J. Proteome Res., 2017, 16, 2101-2112. DOI: 10.1021/acs.jproteome.7b00107.
Article
-
Crowe, S. O.; Rana, A. S.; Deol, K. K.; Ge, Y.; Strieter, E. R.
Ubiquitin Chain Enrichment Middle-Down Mass Spectrometry Enables Characterization of Branched Ubiquitin Chains in Cellulo.
Anal Chem., 2017, 89, 4428-4434. DOI: 10.1021/acs.analchem.6b03675.
Article
-
Gregorich, Z. R.; Cai, W.; Jin, Y.; Wei, L.; Chen, A. J.; McKiernane, S. H.; Aiken, J. M.; Moss, R. L.; Diffee, G. M.; Ge, Y.
Top-down targeted proteomics reveals decrease in myosin regulatory light chain phosphorylation that contributes to sarcopenic muscle dysfunction,
J. Proteome Res., 2016, 15 (8), pp 2706-2716. DOI: 10.1021/acs.jproteome.6b00244.
Article
- Crowe, S.O.; Pham, G.H.; Ziegler, J.C.; Deol, K.K.; Guenette, R.G.; Ge, Y.; Strieter, E.R. Subunit-specific labeling of ubiquitin chains by using sortase: insights into the selectivity of deubiquitinases. ChemBioChem., 2016, 17(16):1525-31. DOI: 10.1002/cbic.201600276. Article
-
Cai, W.; Tucholski, T. M.; Gregorich Z. R.; Ge, Y.
Top-down proteomics: technology advancements and application to heart diseases.
Expert Rev. Proteomics., 2016, 13, 717-730. DOI: 10.1080/14789450.2016.1209414
Article
- Jin, Y.; Peng, Y.; Lin, Z.; Chen, Y. C.; Wei, L.; Hacker, T. A.; Larsson, L.; Ge, Y. Comprehensive analysis of tropomyosin isoforms in skeletal muscles by top-down proteomics. J Muscle Res Cell Motil., 2016, 37(1-2):41-52. DOI: 10.1007/s10974-016-9443-7. Article
-
Hwang, L.; Guardado-Alvarez†, T.; Ayaz-Gunner, S.; Ge, Y.; Jin S.
A Family of Photolabile Nitroveratryl-Based Surfactants That Self-Assemble into Photodegradable Supramolecular Structures.
Langmuir, 2016, 32 (16): pp 3963-3969.
Article
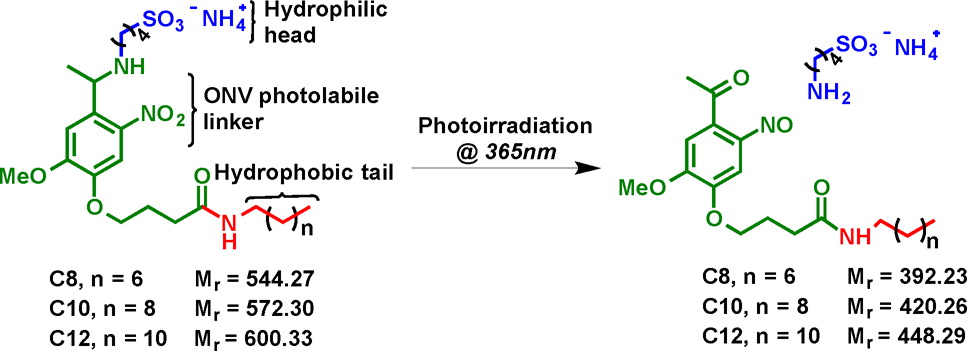
- Chen, B.; Peng, Y.; Valeja, S. G.; Xiu, L.; Alpert, A. J.; Ge, Y.
Online Hydrophobic Interaction Chromatography-Mass spectrometry for Top-down Proteomics.
Anal. Chem. 2016, 88(3):1885-91.
Article
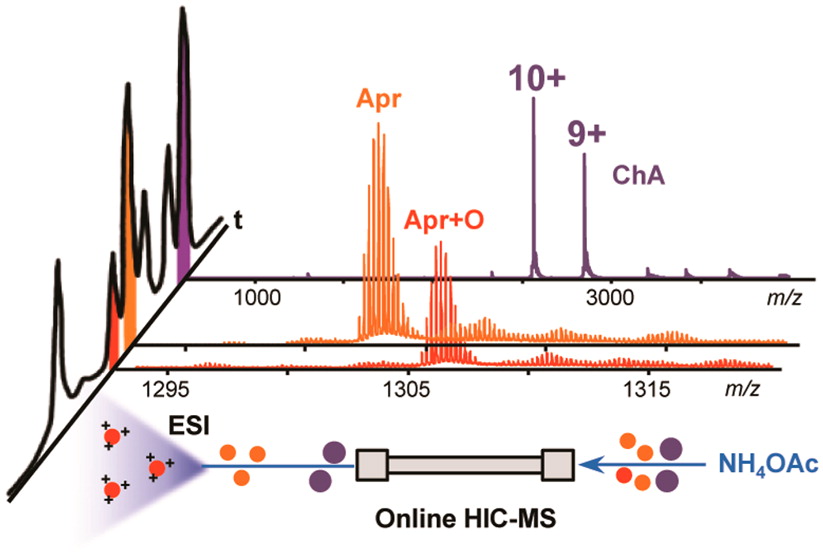
-
Cai, W.; Guner, H.; Gregorich, Z. R.; Chen, A. J.; Ayaz-Guner, S.; Peng, Y.; Valeja, S. G.; Liu, X.; Ge, Y.
MASH Suite Pro: A Comprehensive Software Tool for Top-down Proteomics.
Mol. Cell. Proteomics. 2016, 15(2):703-14.
Article
-
Yu, D.; Peng, Y.; Ayaz-Guner, S.; Gregorich Z. R.; Ge, Y.
Comprehensive characterization of AMP-activated protein kinase catalytic domain by top-down mass spectrometry.
J. Am. Soc. Mass Spectrom. 2016, 27(2): 220-232
Article
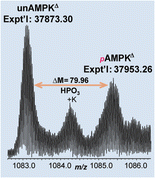
- Gregorich Z. R.; Peng, Y.; Wolff, J. J.; Guo, W.; Guner, H.; Doop, J.; Hacker, T. A.; Ge, Y. Comprehensive assessment of regional and transmural heterogeneity in myofilament protein phosphorylation by top-down mass spectrometry. J. Mol. Cell. Cardiol. 2015, 87, 103-112. Article
-
Chen, Y.-C.; Ayaz-Guner, S.; Peng, Y.; Lane, M. L.; Locher, M.; Larsson, L.; Kohmoto, T.; Moss, R. L.; Ge, Y.
A top-down LC/MS+ method for assessing actin isoforms as a potential cardiac disease marker.
Anal Chem. 2015, 87(16): 8399-406.
Article
-
Chang, Y. H.; Ye, L.; Cai, W.; Lee, Y.; Guner, H.; Lee, Y.; Kamp, T. J.; Zhang, J.; Ge, Y.
Quantitative Proteomics Reveals Differential Regulation of Protein Expression in Recipient Myocardium after Trilineage Cardiovascular Cell Transplantation.
Proteomics. 2015, 15, 2560-2567 DOI: 10.1002/pmic.201500131.
Article
-
Valeja, S. G.; Xiu, L.; Gregorich, Z. R.; Guner, H.; Jin, S.; Ge, Y.
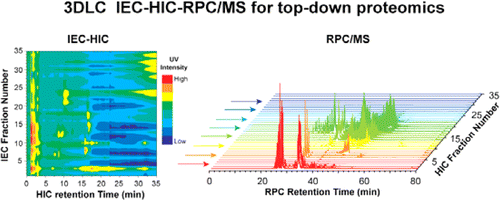
Three dimensional liquid chromatography coupling ion exchange chromatography/hydrophobic interaction chromatography/reverse phase chromatography for effective protein separation in top-down proteomics.
Anal Chem. 2015, 87(10), 5363-71.
Article
-
Hwang, L.; Ayaz-Guner, S.; Cai, W.; Gregorich Z. R.; Jin, S.; Ge, Y.
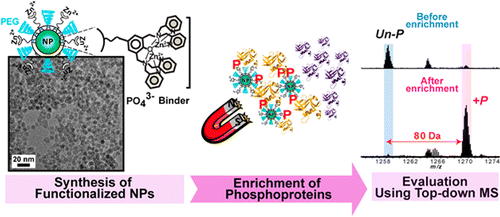
Specific enrichment of phosphoproteins using functionalized multivalent nanoparticles. J. Am. Chem. Soc., 2015, 87, 5363-5371.
Article
-
Chang, Y.; Gregorich Z. R.; Chen, A. J.; Hwang L.; Guner, H.; Yu, D.; Zhang, J.; Ge, Y.
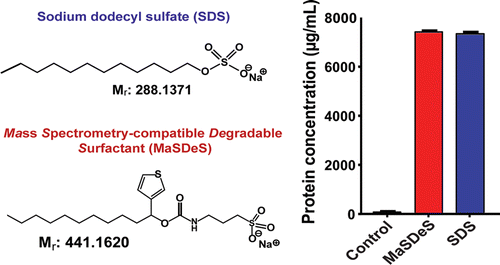
New mass spectrometry-compatible degradable surfactant for tissue proteomics. J. Proteome Res., 2015, 14, 1587-1599.
Article
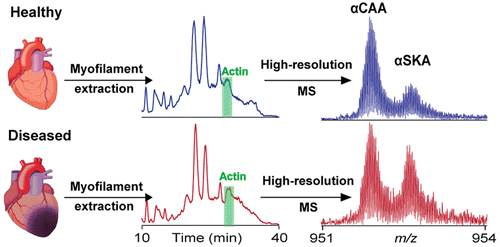
- Chen, Y.; Sumandea, M. P.; Larsson, L.; Moss, R. L.; Ge, Y. Dissecting human skeletal muscle troponin proteoforms by top-down mass spectrometry. J. Muscle Res. Cell Motil., 2015, 36, 168-181. Article
- Ye, L.; Chang, Y. H.; Xiong Q.; Zhang P.; Somasundaram, P.; Lepley M.; Swingen C.; Su, L.; Wendel, J. S.; Guo, J.; Jang, A.;
Rosenbush, D.; Zhang, L.; Greder, L.; Dutton, J. R.; Zhang, J.; Kamp, T. J.; Kaufman, D.S.; Ge, Y.; Zhang, J. Cardiac repair
in a porcine model of acute myocardial infarction with human induced pluripotent stem cell-derived cardiovascular cells.
Cell Stem Cell, 2014, 15, 750-761.
Article
- Peng, Y.; Gregorich Z. R.; Valeja, S. G.; Zhang, H.; Cai, W.; Chen, Y.; Guner, H.; Chen, A. J.; Schwahn, D. J.; Hacker, T. A.; Liu, X.; Ge, Y.
Top-down proteomics reveals concerted reductions in myofilament and Z-disc protein phosphorylation after acute myocardial infarction.
Mol. Cell. Proteomics, 2014, 13, 2752-2764.
Article
- Ge, Y.; Van Eyk, J. Cardiovascular disease: the leap towards translational and clinical proteomics. Proteomics Clin. Appl., 2014, 8, 473-475. Article
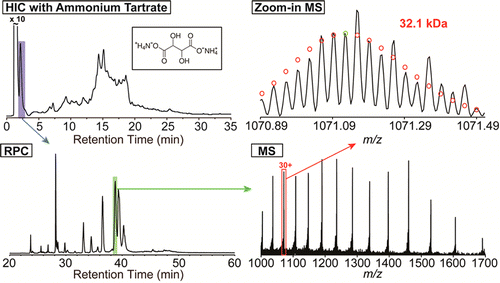
- Xiu, L.; Valeja, S. G.; Alpert, A. J.; Jin, S.; Ge, Y. Effective protein separation by coupling hydrophobic interaction and reverse phase chromatography for top-down proteomics.
Anal. Chem. 2014, 86, 7899-7906.
Article
- Valkevich, E. M.; Sanchez, N.; Ge, Y.; Strieter, E. R. Middle-down mass spectrometry enables characterization of branched ubiquitin chains.
Biochemistry, 2014, 53, 4979−4989.
Article
- Peng, Y.; Ayaz-Guner, S.; Yu, D.; Ge, Y. Top-down mass spectrometry of cardiac myofilament proteins in health and disease, Proteomics Clin. Appl. 2014, 8, 554-568. Article
- Gregorich Z. R.; Ge, Y. Top-down proteomics in health and disease: challenges and opportunities, Proteomics, 2014, 14, 1195-1210. *This is an invited article for a special issue on “Top-down proteomics”. Article
- Gregorich Z. R.; Chang, Y.-W.; Ge, Y. Proteomics in heart failure: top-down or bottom-up? Pflugers Arch. Eur. J. Physiol. 2014, 466, 1199-1209. *This is an invited review for a special issue on “Heart Failure”. Article
- Witayavanitkul, N.; Sarkey, J.; Mou, Y. A.; Kuster, D.W.D.; Khairallah, R. J.; Govindan, S.; Chen, X.; Ge, Y.; Rajan, S.; Wieczorek, D. F.; Irving, T.; de Tombe, P. P.; Sadayappan, S. Myocardial infarction-induced N-terminal fragment of cMyBP-C impairs myofilament function in human left ventricular myofibrils, J. Biol. Chem. 2014, 289, 8818-8827 Article
- Chen, S.; Zhu, P.; Guo, H.; Wang, Y.; Ma, Y.; Wang, J.; Gao, J.; Chen, J.; Ge, Y.; Zhuang, J.; Li, J. Alpha-1 catalytic subunit of AMPK modulates contractile function of cardiomyocytes through phosphorylation of troponin I, Life Sci., 2014, 98, 75-82. Article
- Guner, H.; Close, P.L.; Cai, W.; Zhang, H.; Peng, Y.; Chen, Y.; Gregorich, Z. R. Ge, Y. MASH Suite: A user-friendly and versatile software interface for high-resolution mass spectrometry data interpretation and visualization,
J. Am. Soc. Mass Spectrom. 2014, 25, 464-470.
Article
- Zhao, D. S.; Gregorich Z. R.; Ge, Y. High throughput screening of disulfide-containing proteins in a complex mixture, Proteomics, 2013, 13, 3256-3260. Article
- Peng, Y.; Yu, D.; Gregorich Z., Chen, X.; Beyer, A. M.; Gutterman, D. D.; Ge, Y. In-depth proteomic analysis of human tropomyosin by top-down mass spectrometry, J. Muscle Res. Cell Motil. 2013, 34, 199-210. *This is an invited article for a special edition on “Tropomyosin: form and function”. Article
- Miao, J.; Lawrence, M.; Jeffers, V.; Zhao, F.; Parker, D.; Ge, Y.; Sullivan, W. J.; Cui, L. Extensive lysine acetylation occurs in evolutionarily conserved metabolic pathways and parasite-specific functions during Plasmodium falciparum intraerythrocytic development, Mol. Microbiol. 2013, 89, 660-675. Article
- Chen, X.; Ge, Y. Ultra-high pressure fast size exclusion chromatography for top-down proteomics, Proteomics, 2013, 13, 2563-2566. Article
- Smith LM, Kelleher NL; Consortium for Top Down Proteomics (Linial M, Goodlett D, Langridge-Smith P, Goo YA, Safford G, Bonilla L, Kruppa G, Zubarev R, Rontree J, Chamot-Rooke J, Garavelli J, Heck A, Loo J, Penque D, Hornshaw M, Hendrickson C, Pasa-Tolic L, Borchers C, Chan D, Young N, Agar J, Masselon C, Gross M, McLafferty F, Tsybin Y, Ge Y, Sanders I, Langridge J, Whitelegge J, Marshall A.), Proteoform: a single term describing protein complexity. Nat Methods. 2013, 10, 186-187. Article
- Guy, M. J.; Chen, Y.; Clinton, L.; Zhang, H.; Zhang, J.; Dong, X.; Xu, Q.; Ayaz-Guner, S.; Ge, Y. The impact of antibody selection on the detection of cardiac troponin I, Clin. Chim. Acta. 2013, 420, 82-88. *This is an invited article for a special issue on “Mass Spectrometry for Clinical Diagnosis”. Article
- Peng, Y.; Chen, X.; Zhang, H.; Xu, Q.; Hacker, T. A.; Ge, Y. Top-down targeted proteomics for deep sequencing of tropomyosin isoforms, J. Proteome Res. 2013, 12, 187-198. Article
- Trang, V. H.; Valkevich, E. M.; Minami, S.; Chen, Y.; Ge, Y.; Strieter, E. R. Nonenzymatic polymerization of ubiquitin: single-step synthesis and isolation of discrete ubiquitin oligomers, Angew. Chem. Int. Ed. 2012, 124, 13262-13265. Article
- Yang, C.; Park, A. C.; Davis, N. A,; Russell, J. D.; Kim, B.; Brand, D. D.; Lawrence, M. J.; Ge, Y.; Coon, J. J.; Greenspan, D. S.; Comprehensive mass spectrometric mapping of the hydroxylated amino acid residues of the a1(V) collagen chain, J. Biol. Chem. 2012, 287, 40598-40610 Article
- Jia, C.; Hui, L.; Cao, W.; Lietz, C. B.; Jiang, X.; Chen, R.; Catherman, A. D.; Thomas, P. M.; Ge, Y.; Kelleher, N. L.; Li.; L. High-definition de novo sequencing of crustacean hyperglycemic hormone (CHH)-family neuropeptides, Mol. Cell Proteomics, 2012, 11, 1951-64. Article
- Ge, Y.; Moss, R. L. Nitroxyl, redox switches, cardiac myofilaments, and heart failure: A prequel to novel therapeutics? Circ. Res. 2012, 111, 954-956. Article
- Peng, Y.; Chen, X.; Sato, T., Rankin, S. A.; Tsuji, R. F.; Ge, Y. Rapid purification and high-resolution top-down mass spectrometric characterization of human salivary α-amylase, Anal. Chem. 2012, 84, 3339-3346. Article
- Oliveira, S.M.; Zhang, Y.H.; Sancho Solís, R.; Isackson, H.; Bellahcene, M.; Yavari, A.; Pinter, K.; Davies, J.D., Ge, Y.; Ashrafian, H.; Walker, J. W.; Carling, D.; Watkins, H.; Casadei, B.; Redwood, C. AMP-activated protein kinase phosphorylates cardiac troponin I and alters contractility of murine ventricular myocytes, Circ. Res., 2012, 110, 1192-1201.
- Valkevich, E. M.; Guenette, R. G.; Sanchez, N. A.; Chen,
Y.; Ge, Y.; Strieter, E. R. Forging isopeptide bonds using
thiol-ene chemistry: site-specific coupling of ubiquitin
molecules for studying the activity of isopeptidases, J. Am.
Chem. Soc. 2012, 134, 6916-6919.
- Dong, X.; Sumandea, C. A.; Chen, Y.; Garcia-Cazarin, M. L.; Zhang, J.; Balke, C. M.; Sumandea, M. P.; Ge, Y. Augmented phosphorylation of cardiac troponin I in hypertensive heart failure, J. Biol. Chem. 2012, 287, 848-857.
- Du, J.; Teng, R. J.; Lawrence M.; Guan, T.; Ge, Y.; Shi, Y. The protein partners of GTP cyclohydrolase I (GCH1) in rat organs, Plos One, 2012, 7, e33991.
- Wang, Y.; Pinto, J. R.; Sancho Solis, R.; Liang, J.; Diaz-Perez, Z.; Harada, K.; Ge, Y.; Walker, J. W. Potter, J. D. The generation and functional characterization of knock in mice harboring the cardiac Troponin I-R21C mutation associated with hypertrophic cardiomyopathy, J. Biol. Chem. 2012, 287, 2156-2167.
- Zhang, H.; Ge, Y. Comprehensive analysis of protein modifications by top-down mass spectrometry, Circ. Cardiovasc. Genet. 2011, 4, 711. *This is an invited review for “Integrating proteomics into cardiovascular disease - an educational review series”.
- Zhang, J.; Guy, J. M.; Norman, H. A.; Chen, Y.; Dong, X.; Wang, S.; Kohmoto, T.; Young, K. H.; Moss, R. L.; Ge, Y. Top-Down quantitative proteomics identified phosphorylation of cardiac troponin I as a candidate biomarker for chronic heart failure, J. Proteome Res. 2011, 10, 4054-4065.
- Zhang, J.; Zhang, H.; Ayaz-Guner, S.; Dong, X.; Xu, Q.; Guner, H.; Ge, Y. Phosphorylation, but not alternative splicing or proteolytic degradation, is conserved in human and mouse cardiac troponin T, Biochemistry, 2011, 50, 6081-6092.
- Sancho Solis, R.; 41. Guy, M. J.; Chen, Y.; Clinton, L.; Zhang, H.; Zhang, J.; Dong, X.; Xu, Q.; Ayaz-Guner, S.; Ge, Y. The impact of antibody selection on the detection of cardiac troponin I, Clin. Chim. Acta. Accepted. *This is an invited article.
- Kuhn, P.; Chumanov, R.; Wang, Y.; Ge, Y.; Burgess, R. R.; Xu, W. Automethylation of CARM1 allows coupling of transcription and mRNA splicing, Nucl. Acids Res. 2011, 39, 2717-2726.
- Xu, F.; Xu, Q.; Dong, X.; Guy, M.; Guner, H.; Hacker, T. A.; Ge, Y. Top-down high-resolution electron capture dissociation mass spectrometry for comprehensive characterization of post-translational modifications in rhesus monkey cardiac troponin I, Int. J. Mass Spectrom. 2011, 305, 95-102. *This is an invited article.
- Nelson, C. A.; Szezech, J. R.; Dooley, C. J.; Xu, Q.; Lawrence, M. J.; Zhu, R.; Jin, S.; Ge, Y. Effective enrichment and mass spectrometry analysis of phosphopeptides using mesoporous metal oxide nanomaterials, Anal. Chem. 2010, 82, 7193-7201.
- Zhang, J.; Dong, X.; Hacker, T. A.; Ge, Y. Deciphering modifications in swine cardiac troponin I by Top-down high-resolution tandem mass spectrometry, J. Am. Soc. Mass. Spectrom. 2010, 21, 940-948. * This is an invited article featured on the cover of the journal.
- Ayaz-Guner, S.; Zhang, J.; Li, L.; Walker, J. W.; Ge, Y. In vivo phosphorylation site mapping in mouse cardiac troponin I by high resolution top-down electron capture dissociation mass spectrometry: Ser22/23 are the only sites basally phosphorylated, Biochemistry, 2009, 48, 8161-8170.
- Ge, Y.; Rybakova, I.; Xu, Q.; Moss, R. L. Top-down high resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state, Proc. Natl. Acad. Sci. U. S. A. 2009,106, 12658-12663. *This article is a PNAS Direct Submission (Track II).
- Kuhn, P.; Xu, Q.; Cline, E.; Zhang, D.; Ge, Y.; Xu, W. Delineating Anopheles gambiae coactivator associated arginine methyltransferase 1 (AgCARM1) by top-down high resolution tandem mass spectrometry, Protein Sci. 2009, 18, 1272-1280.
- Du, J.; Wei, N; Xu, H.; Ge, Y.; Vasquez-Vivar, J.; Guan, T.; Oldham, K. T.; Pritchard, K. A.; Shi, Y. Identification and functional characterization of phosphorylation sites on GTP cyclohydrolase I, Arterioscler. Thromb. Vasc. Biol. 2009, 29, 2161-2168.
- Nelson, C. A.; Szczech, J. R.; Xu, Q.; Jin, S.; Ge, Y. Mesoporous Zirconium oxide nanomaterials effectively enrich phosphopeptides for mass spectrometry-based phosphoproteomics, Chem. Commun. 2009, 6607-6609.
- Liu, L.; Hou, J.; Du, J.; Chumanov, R. S.; Xu, Q.; Ge, Y.; Johnson, J.A.; Murphy, R.M. Differential modification of Cys10 alters transthyretin’s effect on beta-amyloid aggregation and toxicity, Protein Eng. Des. Sel. 2009, 1-10.
- Ma, M.; Chen, R.; Ge, Y.; He, H; Marshall, A. G.; Li, L. Combining bottom-up and top-down mass spectrometric strategies for de novo sequencing of the Crustacean hyperglycemic hormone (CHH) from Cancer borealis, Anal. Chem. 2009, 81, 240-247.
- Sancho Solis, R.; Ge, Y.; Walker, J. W. Single amino acid sequence polymorphisms in rat cardiac troponin revealed by top-down tandem mass spectrometry, J. Muscle Res. Cell Motil. 2008, 29, 203-212.
- Zabrouskov, V.; Ge, Y.; Schwartz, J.; Walker, J. W. Unraveling molecular complexity of phosphorylated human cardiac troponin I by top down electron capture dissociation/electron transfer dissociation mass spectrometry, Mol. Cell. Proteomics 2008, 7, 1838-1849.
2025
For more recent updates, please see https://scholar.google.com/citations?hl=en&user=Ymgpd5QAAAAJ&view_op=list_works&sortby=pubdate
2024
2023
2022
2021
2020
2019
2018
2017
2016
2015
2014
2013
2012
2011
2010
2009
2008
Prior to UW-Madison:
2001-2005- Colabroy, K. L.; Zhai, H. L.; Li, T. F.; Ge, Y.; Zhang, Y.; Liu, A. M.; Ealick, S. E.; McLafferty, F. W.; Begley, T. P., The mechanism of inactivation of 3-hydroxyanthranilate-3,4-dioxygenase by 4-chloro-3-hydroxyanthranilate. Biochemistry 2005, 44, 7623-7631.
- Xiang, J.; Ipek, M.; Suri, V.; Massefski, W.; Pan, N.; Ge, Y.; Tam, M.; Xing, Y.; Tobin, J.; Xu, X.; Tam, S. Synthesis and biological evaluation of sulfonamidooxazoles and b-keto sulfones: selective inhibitors of 11b-hydroxyster oid dehydrogenase type I, Bioorg. Med. Chem. Lett. 2005, 15, 2865-2869.
- Ge, Y.; Lawhorn, B. G.; ElNaggar, M.; Sze, S. K.; Begley, T. P.; McLafferty, F. W. Detection of four oxidation sites in viral prolyl-4-hydroxylase by top-down mass spectrometry, Protein Sci. 2003, 12, 2320-2326.
- Ge, Y.; ElNaggar, M.; Sze, N.; Oh, H.; Begley, T. P.; McLafferty, F. W.; Boshoff, H.; Barry, C. B. Top down characterization of secreted proteins from Mycobacterium tuberculosis by electron capture dissociation mass spectrometry. J. Am. Soc. Mass Spectrom. 2003, 14, 253-261.
- Sze, S. K.; Ge, Y.; Oh, H.; McLafferty, F. W. Plasma electron capture dissociation for the characterization of large proteins by top down mass spectrometry Anal. Chem. 2003, 75, 1599-1603.
- Oh, H.; Bruker, K.; Sze, S. K.; Ge, Y.; McLafferty, F. W. Secondary and tertiary structure of gaseous ions characterized by electron capture dissociation mass spectrometry and photofragment spectroscopy, Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 15863-15868.
- Sze, S. K.; Ge, Y.; Oh, H.; McLafferty, F. W. Top down mass spectrometry of a 29 kDa protein for characterization of any posttranslational modification to within one residue, Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 1774-1779.
- Ge, Y.; Lawhorn, B. G.; ElNaggar, M.; Strauss, E.; Park, J-H.; Begley, T. P.; McLafferty, F. W. Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry, J. Am. Chem. Soc. 2002, 124, 672-678
- Xi, J.; Ge, Y.; Kinsland, C. L.; McLafferty, F. W.; Begley, T. P. Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: identification of an acyldisulfide-linked protein--protein conjugate that is functionally analogous to the ubiquitin/E1 complex, Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 8513-8518.
- Ge, Y.; Horn, D. M.; McLafferty, F. W. Blackbody infrared radiative dissociation of larger (42 kDa) multiply charged proteins, Int. J. Mass Spectrom. 2001, 210/211, 203-214.
- McLafferty, F. W.; Horn, D. M.; Ge, Y..; Cerda, B.; Fridriksson, E.; Breuker, K. New frontiers in molecular mass spectrometry, Adv. Mass Spectrom. 2001, 67, 167-173.
- McLafferty, F. W.; Horn, D. M.; Breuker, K.; Ge, Y.; Cerda, B., A. Electron capture dissociation of gaseous multiply charged ions by Fourier-transform ion cyclotron resonance, J. Am. Soc. Mass Spectrom. 2001, 12, 245-249.
- Strauss, E.; Kinsland, C.; Ge, Y.; McLafferty, F. W.; Begley, T. P. Phosphopantothenoylcysteine synthetase from Escherichia coli: Identification and characterization of the last unidentified coenzyme A biosynthetic enzyme in bacteria. J. Biol. Chem. 2001, 276, 13513-13516.
- Horn, D. M.; Ge, Y.; McLafferty, F. W. Activated ion electron capture dissociation for mass spectral sequencing for larger (42 kDa) proteins, Anal. Chem. 2000, 72, 4778-4784.
