Cardiovascular Systems Biology
Through High-resolution Mass Spectrometry-based Comparative Proteomics and Metabolomics
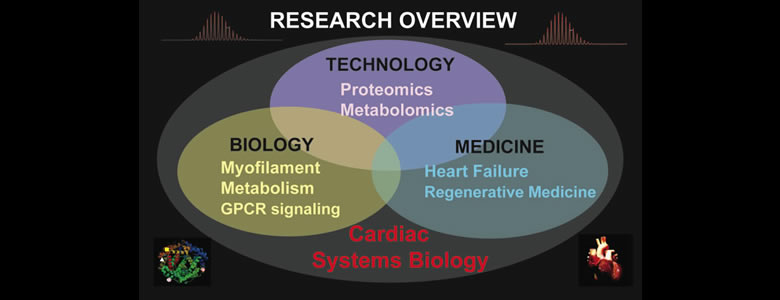
Research Overview
Our research aims to understand the molecular and cellular mechanisms underlying cardiovascular diseases via systems biology approaches featuring cutting-edge high-resolution mass spectrometry (MS)-based comparative proteomics and metabolomics in conjunction with functional studies. Our research is highly interdisciplinary within the interface of chemistry, biology, and medicine. Success in my proposed research will advance our understanding of the molecular basis of diseases and foster the development of new strategies for early diagnosis, prevention and better treatment of cardiovascular diseases. It is our belief that in order to make significant impact in molecular medicine, we need to combine technological advances with functional studies and bridge the silos between chemical and biological sciences as well as basic and translational/clinical research.
Cardiovascular disease is the leading cause of morbidity and mortality in developed countries and is reaching epidemic proportions. Transformative insights from a holistic approach at the systems level have great potential to elucidate disease mechanisms and to develop new therapeutic treatments. Proteins and metabolites are important molecular entities of the cell downstream of genes. Hence in the post genomic era, proteomics and metabolomics (the large-scale global analysis of proteins and metabolites in a cell, organism, tissue, and biofluid), are essential for deciphering how molecules interact as a system and for understanding the functions of cellular systems in health and disease. However, there are tremendous challenges in proteomics and metabolomics due to the extreme complexity and dynamic nature of the proteome and metabolome.
To address such challenges, we are developing
novel ultra high-resolution MS-based top-down comparative
proteomics and metabolomics platforms for systems biology studies
with high efficiency, specificity, sensitivity, and
reproducibility. Specifically we are focusing on the following
technology developments:
1) Top-down proteomics: We are establishing a top-down disease proteomics platform to provide a comprehensive tool for separation, detection, identification, quantitation and characterization of intact proteins extracted from tissue samples to reveal all disease-related changes in protein expression and post-translational modifications (PTMs) with high efficiency and reproducibility. Particularly, we are developing novel technologies to address the current challenges in top-down proteomics including protein solubility, proteome complexity, and dynamic range. [Read More]
2)
Nanoproteomics: Combining the advances in nanotechnology and
proteomics, we are developing “smart” antibody-like multivalent
nanomaterials to enrich low abundance proteins or PTMs. A
current focus is to develop multivalent nanoparticle reagents for
capturing phosphoproteins globally out of human proteome followed
by top-down MS analysis of intact phosphoproteins.
3)
Metabolomics: We are establishing an integrated ultra
high-resolution MS-based platform for rapid, sensitive,
reproducible detection and accurate quantification of diverse
metabolites over a wide range of concentrations in a
high-throughput and automatic fashion.
We then
employ these technology platforms to study heart failure in
conjunction with functional assays. Currently we are focused
on two major directions:
i) Cardiac myofilaments: We are testing
the hypothesis that both extrinsic and intrinsic stresses trigger
the molecular signaling processes that could result in altered
modifications to myofilaments leading to contractile dysfunction
and eventually failure. We employ an unbiased top-down proteomics
analysis of the myofilaments to globally detect the changes in
protein modifications, identify which sites are modified or
altered, and elucidate how these alterations act in concert during
the transition to the heart failure. We then mechanistically
link the proteomic changes to in vitro, ex vivo, and in vivo
cardiac function to assess synergistic changes of myofilaments in
regulating contractility during left ventricular remodeling to
heart failure using large animal (swine) models and human clinical
studies.
ii) Cardiac regeneration: We are testing the
hypothesis that transplanted stem cells can trigger the molecular
signaling pathways which would protect the native cardiomyocytes
from apoptosis and reverse pathological left ventricular
remodeling. We aim to gain a holistic view of the molecular
signaling pathways underlying cardiac regeneration in response to
stem cell therapy through novel systems biology approaches.
Our Research Statement [Link]
Currently funded research projects
"Top-Down
Proteomics of Myofilaments in Heart Failure"
Principal Investigator: Ge
Agency: NIH/NHLBI
Type: R01, R01HL096971
Period: 8/5/2011 - 11/30/2022
Goals: o develop top-down mass spectrometry-based proteomics technologies for analysis of key myofilament regulatory proteins and to understand the disease mechanism in left ventricular hypertrophy and failure using a pressure overload animal model and hypertrophic cardiomyopathy. A multi-disciplinary approach featuring top-down proteomics, induced pluripotent stem cells derived cardiomyocytes (iPSC-CMs), and cardiac tissue engineering will be used to delineate the underlying mechanisms of hypertrophic cardiomyopathy.
[Read More]
“Deciphering Myofilament Modifications in Ischemic Cardiomyopathy”
Principal Investigator: Ge
Agency: NIH
Type: R01, R01HL10980
Period: 3/1/2013-6/30/2025
Goals: The major goal of this proposal is to delineate the molecular mechanism underlying early-phase post-myocardial infarction (MI) ventricular remodeling through a novel multi-omics approach. The most recent renewal has three aims: Aim 1. Identify concerted changes in the myofilament and Z-disc proteins in post-MI swine myocardium and relate to contractile dysfunction in both sexes. Aim 2. Determine the metabolic alterations in both proteome and metabolome in the post-MI remodeling. Aim 3. Identify sarcomeric and metabolic markers in ischemic cardiomyopathy (ICM) patient myocardium and assess their functional consequences in consideration of comorbidities and sex differences.
“MASH Explorer, a Comprehensive Software Environment of Top-Down Proteomics”
Principal
Investigator: Ge
Agency: NIH/NIGMS
Type: R01, R01GM125085
Period: 6/01/2018 - 3/31/2022
Goals: To develop MASH Explorer, a comprehensive, user-friendly, and universal software environment for top-down proteomics, to process data from various vendor formats and incorporate multiple algorithms for deconvolution and database search with user-friendly graphical interfaces.
[Read More]
“Enabling Top-down Proteomics through Material Chemistry and Nanotechnology”
Principal Investigator: Ge and Jin (MPI)
Agency: NIH/NIGMS
Type: R01, R01GM117058
Period: 9/22/2015-6/30/2024
Goals: To develop novel approaches enabled by nanotechnology and materials chemistry to address the challenges in top-down MS-based proteomics.
[Read More]
“KCNJ2 Mutation-Induced Arrhythmia Mechanisms in CPVT Phenotypes”
Principal Investigator:Eckhardt (Co-Investigator: Ge)
Agency: NIH/NHLBI
Type: R01, R01HL139738
Period: 8/1/2018 – 7/31/2022
Goals: To determine the biophysical properties, Ca2+ sensitivity, phosphorylation state and arrhythmia mechanism of KCNJ2 (ion channel Kir2.1) mutations associated with a catecholaminergic polymorphic ventricular tachycardia or an Adersen-Tawil syndrome phenotype and compare that to a heart failure model.
[Read More]
Sponsors
 |
 |
 |
 
|
Instruments
Mass Spectrometer
 |
 |
LTQ (Thermo Scientific) |
7-Tesla FT-ICR (Thermo Scientific) |
 |
 |
Impact II (Bruker Daltonics) |
Maxis II ETD (Bruker Daltonics) |
 |
|
solariX 12T FTMS (Bruker Daltonics) |
HPLC
 |
 |
Acquity H-Class System (Waters) |
Acquity 2D M-Class System (Waters) |
 |
|
nanoAcquity (Waters) |
