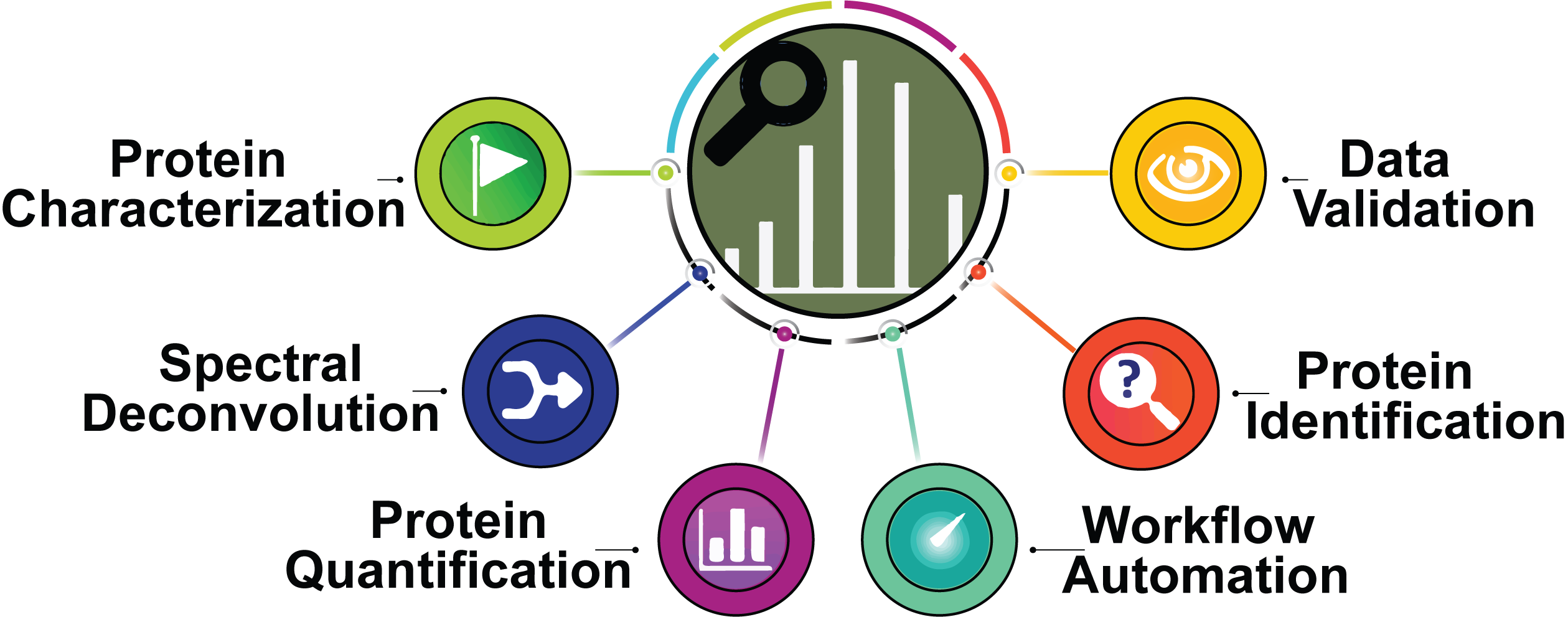
The MASH software series is a freely available collection of data analysis tool for both native and denatured top-down proteomics. Beginning with MASH Suite then MASH Suite Pro, NIH support enabled the release of MASH Explorer, a comprehensive and user-friendly tool for denatured top-down proteomics. MASH Explorer can process data from various vendors and incorporates multiple algorithms for deconvolution and database searching. Flexible data reporting options allow easy data interaction through an intuitive graphical user interface.
The latest offering in the MASH series, MASH Native, is an all-in-one software for native top-down proteomics, complex-down analysis, native intact MS, and denatured top-down. The downloadable MASH Native App is a "one-stop-shop", capable of processing data from various vendor-specific and non-specific formats, and incorporating multiple algorithms for deconvolution and database searching. It is developed by an interdisciplinary team consisting of experts in top-down mass spectrometry-based proteomics, computational data science, bioinformatics and biostatistics. For full functionality, MASH Native is recommended.
The Ge lab would like to acknowledge funding from NIH R01 grant (GM125085) to allow the continuous development of MASH Explorer and MASH Native.
 Learn More |
Raw File Formats Include:
Thermo (*.raw) data files Ms-Deconv MS-Align |